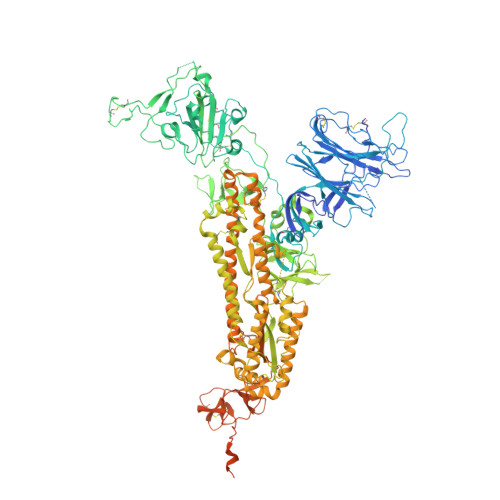
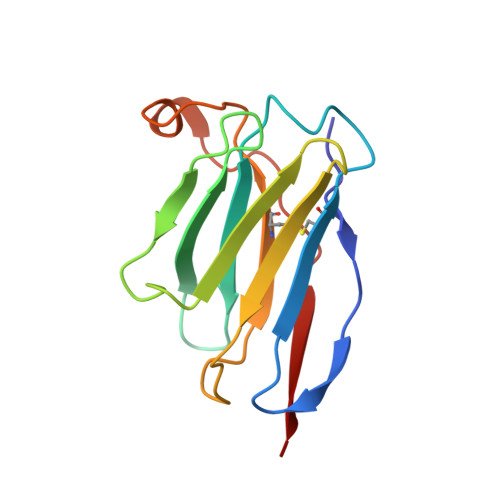
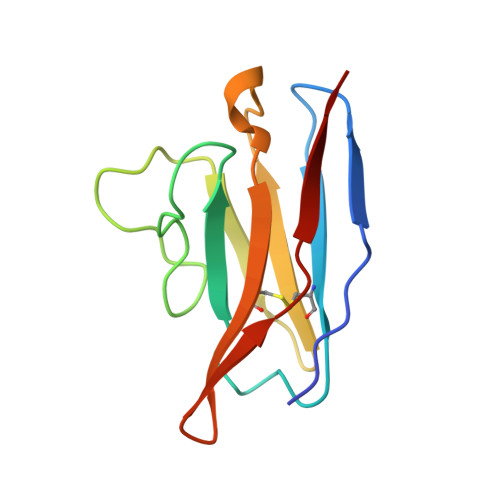
Potent SARS-CoV-2 Neutralizing Antibodies Directed Against Spike N-Terminal Domain Target a Single Supersite
Cerutti, G., Guo, Y., ZHou, T., Gorman, J., Lee, M., Rapp, M., Reddem, E.R., Yu, J., Bahna, F., Bimela, J., Huang, Y., Katsamba, PS., Liu, L., Nair, M.S., Rawi, R., Olia, A.S., Wang, P., Zhang, B., Chuang, G.Y., Ho, D.D., Sheng, Z., Kwong, P.D., Shapiro, L.(2021) Cell Host Microbe