Self-Masked Aldehyde Inhibitors: A Novel Strategy for Inhibiting Cysteine Proteases.
Li, L., Chenna, B.C., Yang, K.S., Cole, T.R., Goodall, Z.T., Giardini, M., Moghadamchargari, Z., Hernandez, E.A., Gomez, J., Calvet, C.M., Bernatchez, J.A., Mellott, D.M., Zhu, J., Rademacher, A., Thomas, D., Blankenship, L.R., Drelich, A., Laganowsky, A., Tseng, C.K., Liu, W.R., Wand, A.J., Cruz-Reyes, J., Siqueira-Neto, J.L., Meek, T.D.(2021) J Med Chem 64: 11267-11287
- PubMed: 34288674
- DOI: https://doi.org/10.1021/acs.jmedchem.1c00628
- Primary Citation of Related Structures:
7M2P - PubMed Abstract:
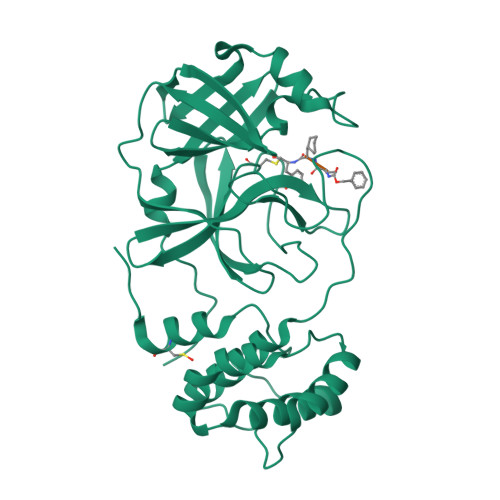
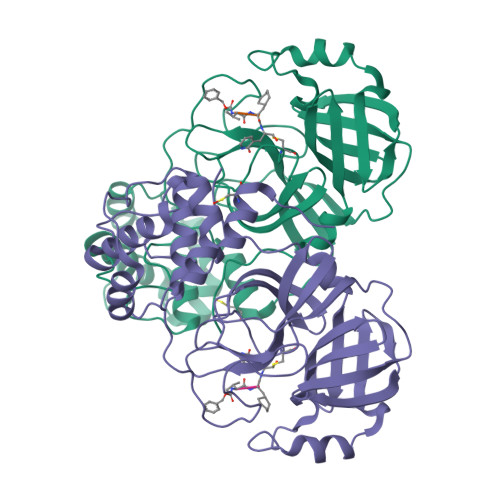
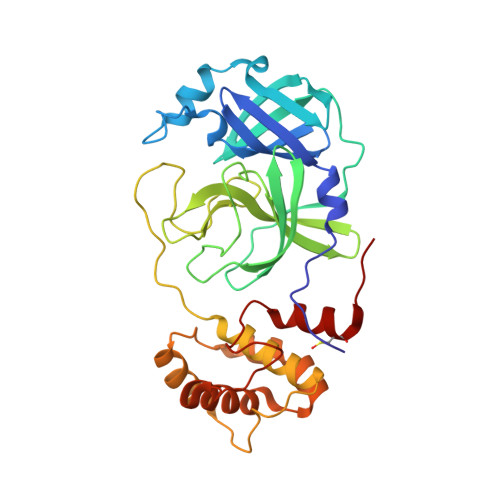
Cysteine proteases comprise an important class of drug targets, especially for infectious diseases such as Chagas disease (cruzain) and COVID-19 (3CL protease, cathepsin L). Peptide aldehydes have proven to be potent inhibitors for all of these proteases. However, the intrinsic, high electrophilicity of the aldehyde group is associated with safety concerns and metabolic instability, limiting the use of aldehyde inhibitors as drugs. We have developed a novel class of self-masked aldehyde inhibitors (SMAIs) for cruzain, the major cysteine protease of the causative agent of Chagas disease- Trypanosoma cruzi . These SMAIs exerted potent, reversible inhibition of cruzain ( K i * = 18-350 nM) while apparently protecting the free aldehyde in cell-based assays. We synthesized prodrugs of the SMAIs that could potentially improve their pharmacokinetic properties. We also elucidated the kinetic and chemical mechanism of SMAIs and applied this strategy to the design of anti-SARS-CoV-2 inhibitors.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Texas A&M University, 300 Olsen Blvd, College Station, Texas 77843, United States.