TRIM5alpha Restriction of HIV-1-N74D Viruses in Lymphocytes Is Caused by a Loss of Cyclophilin A Protection.
Selyutina, A., Simons, L.M., Kirby, K.A., Bulnes-Ramos, A., Hu, P., Sarafianos, S.G., Hultquist, J.F., Diaz-Griffero, F.(2022) Viruses 14: 363
- PubMed: 35215956
- DOI: https://doi.org/10.3390/v14020363
- Primary Citation of Related Structures:
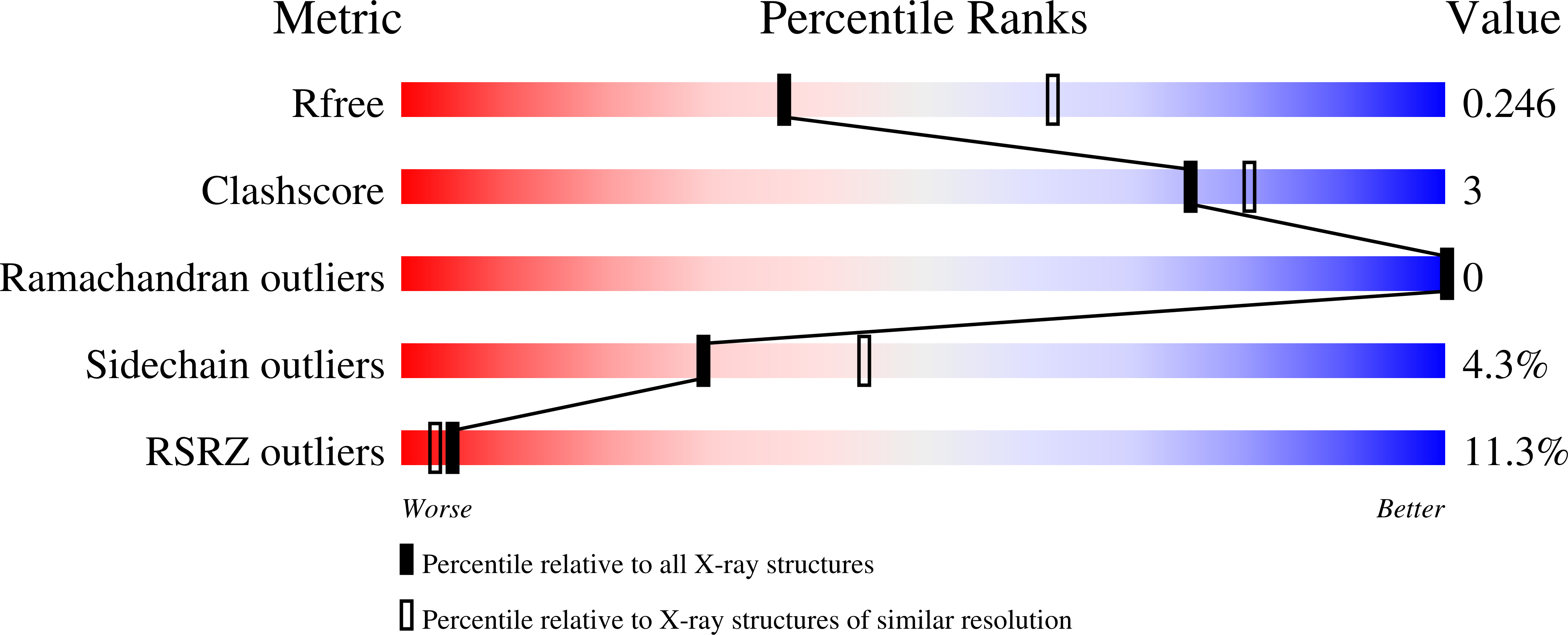
7MKC, 7MN0 - PubMed Abstract:
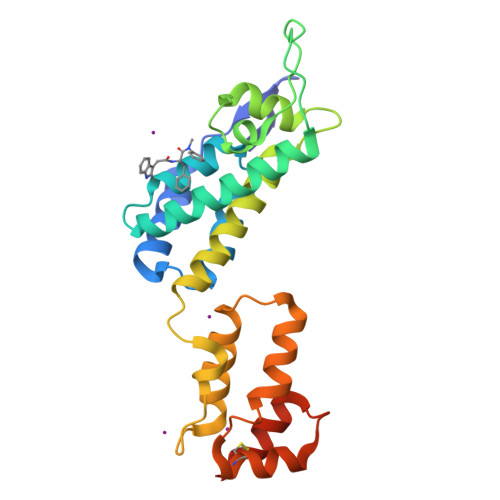
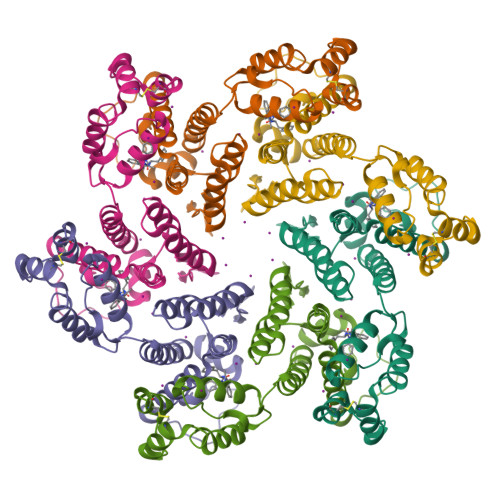
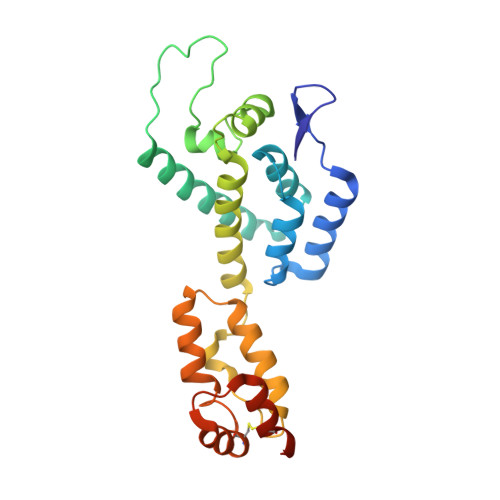
The core of HIV-1 viruses bearing the capsid change N74D (HIV-1-N74D) do not bind the human protein CPSF6. In primary human CD4 + T cells, HIV-1-N74D viruses exhibit an infectivity defect when compared to wild-type. We first investigated whether loss of CPSF6 binding accounts for the loss of infectivity. Depletion of CPSF6 in human CD4 + T cells did not affect the early stages of wild-type HIV-1 replication, suggesting that defective infectivity in the case of HIV-1-N74D viruses is not due to the loss of CPSF6 binding. Based on our previous result that cyclophilin A (Cyp A) protected HIV-1 from human tripartite motif-containing protein 5α (TRIM5α hu ) restriction in CD4 + T cells, we found that depletion of TRIM5α hu in CD4 + T cells rescued the infectivity of HIV-1-N74D, suggesting that HIV-1-N74D cores interacted with TRIM5α hu . Accordingly, TRIM5α hu binding to HIV-1-N74D cores was increased compared with that of wild-type cores, and consistently, HIV-1-N74D cores lost their ability to bind Cyp A. In agreement with the notion that N74D capsids are defective in their ability to bind Cyp A, we found that HIV-1-N74D viruses were 20-fold less sensitive to TRIMCyp restriction when compared to wild-type viruses in OMK cells. Structural analysis revealed that N74D hexameric capsid protein in complex with PF74 is different from wild-type hexameric capsid protein in complex with PF74, which explains the defect of N74D capsids to interact with Cyp A. In conclusion, we showed that the decreased infectivity of HIV-1-N74D in CD4 + T cells is due to a loss of Cyp A protection from TRIM5α hu restriction activity.
Organizational Affiliation:
Department of Microbiology and Immunology, Albert Einstein College of Medicine, Bronx, NY 10461, USA.