Fusion Peptide of SARS-CoV-2 Spike Rearranges into a Wedge Inserted in Bilayered Micelles.
Koppisetti, R.K., Fulcher, Y.G., Van Doren, S.R.(2021) J Am Chem Soc 143: 13205-13211
- PubMed: 34375093
- DOI: https://doi.org/10.1021/jacs.1c05435
- Primary Citation of Related Structures:
7MY8 - PubMed Abstract:
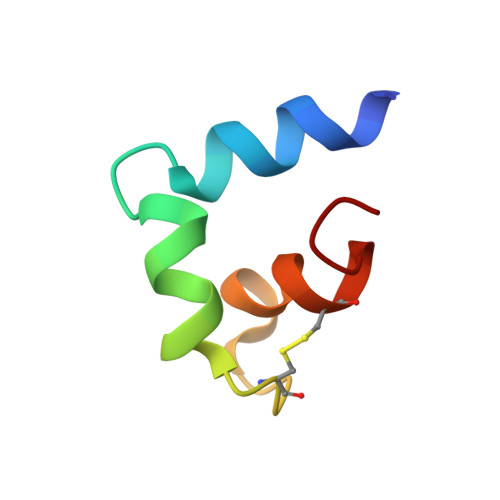
The receptor binding and proteolysis of Spike of SARS-CoV-2 release its S 2 subunit to rearrange and catalyze viral-cell fusion. This deploys the fusion peptide for insertion into the cell membranes targeted. We show that this fusion peptide transforms from intrinsic disorder in solution into a wedge-shaped structure inserted in bilayered micelles, according to chemical shifts, 15 N NMR relaxation, and NOEs. The globular fold of three helices contrasts the open, extended forms of this region observed in the electron density of compact prefusion states. In the hydrophobic, narrow end of the wedge, helices 1 and 2 contact the fatty acyl chains of phospholipids, according to NOEs and proximity to a nitroxide spin label deep in the membrane mimic. The polar end of the wedge may engage and displace lipid head groups and bind Ca 2+ ions for membrane fusion. Polar helix 3 protrudes from the bilayer where it might be accessible to antibodies.
Organizational Affiliation:
Department of Biochemistry, University of Missouri, Columbia, Missouri 65211 United States.