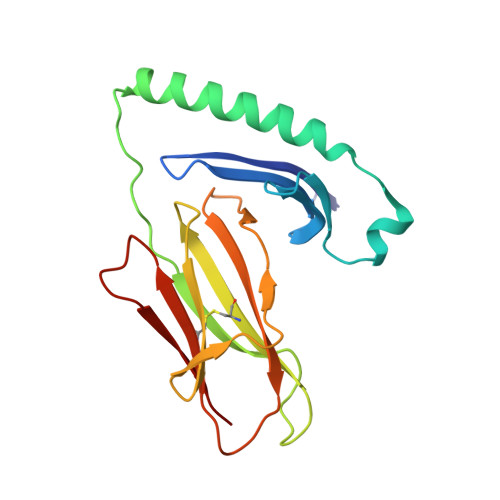
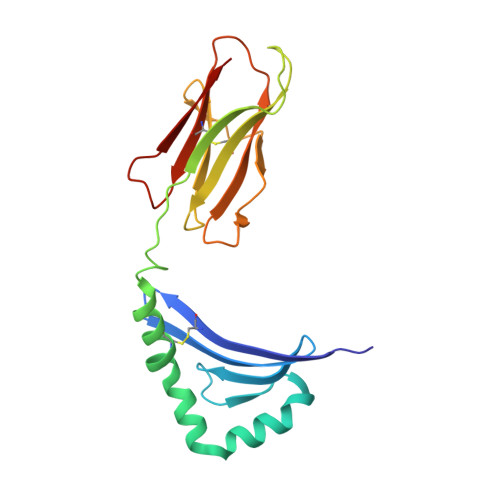
CD4+ T cells in the lungs of acute sarcoidosis patients recognize an Aspergillus nidulans epitope.
Greaves, S.A., Ravindran, A., Santos, R.G., Chen, L., Falta, M.T., Wang, Y., Mitchell, A.M., Atif, S.M., Mack, D.G., Tinega, A.N., Maier, L.A., Dai, S., Pinilla, C., Grunewald, J., Fontenot, A.P.(2021) J Exp Med 218
- PubMed: 34410304
- DOI: https://doi.org/10.1084/jem.20210785
- Primary Citation of Related Structures:
7N19 - PubMed Abstract:
Löfgren's syndrome (LS) is an acute form of sarcoidosis characterized by a genetic association with HLA-DRB1*03 (HLA-DR3) and an accumulation of CD4+ T cells of unknown specificity in the bronchoalveolar lavage (BAL). Here, we screened related LS-specific TCRs for antigen specificity and identified a peptide derived from NAD-dependent histone deacetylase hst4 (NDPD) of Aspergillus nidulans that stimulated these CD4+ T cells in an HLA-DR3-restricted manner. Using ELISPOT analysis, a greater number of IFN-γ- and IL-2-secreting T cells in the BAL of DR3+ LS subjects compared with DR3+ control subjects was observed in response to the NDPD peptide. Finally, increased IgG antibody responses to A. nidulans NDPD were detected in the serum of DR3+ LS subjects. Thus, our findings identify a ligand for CD4+ T cells derived from the lungs of LS patients and suggest a role of A. nidulans in the etiology of LS.
Organizational Affiliation:
Department of Medicine, University of Colorado Anschutz Medical Campus, Aurora, CO.