SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern.
McCallum, M., Bassi, J., De Marco, A., Chen, A., Walls, A.C., Di Iulio, J., Tortorici, M.A., Navarro, M.J., Silacci-Fregni, C., Saliba, C., Sprouse, K.R., Agostini, M., Pinto, D., Culap, K., Bianchi, S., Jaconi, S., Cameroni, E., Bowen, J.E., Tilles, S.W., Pizzuto, M.S., Guastalla, S.B., Bona, G., Pellanda, A.F., Garzoni, C., Van Voorhis, W.C., Rosen, L.E., Snell, G., Telenti, A., Virgin, H.W., Piccoli, L., Corti, D., Veesler, D.(2021) Science 373: 648-654
- PubMed: 34210893
- DOI: https://doi.org/10.1126/science.abi7994
- Primary Citation of Related Structures:
7N8H, 7N8I - PubMed Abstract:
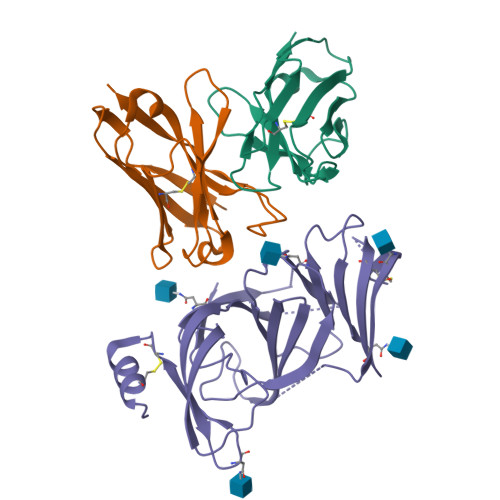
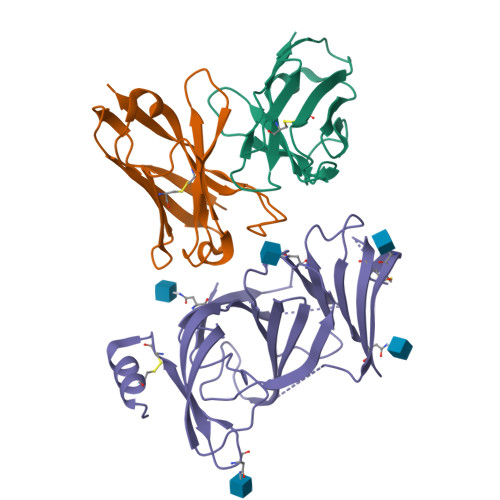
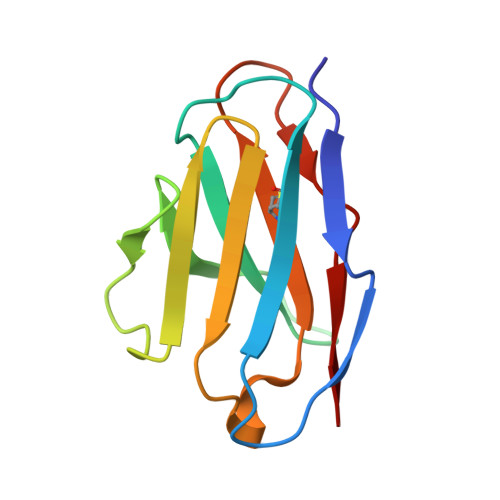
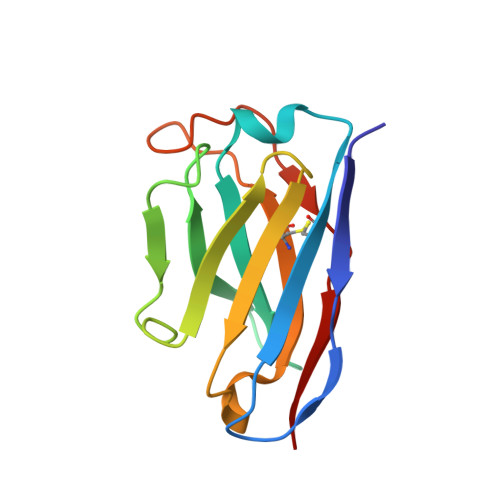
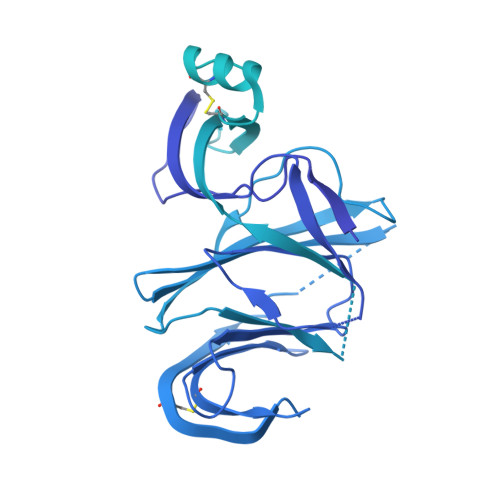
A novel variant of concern (VOC) named CAL.20C (B.1.427/B.1.429), which was originally detected in California, carries spike glycoprotein mutations S13I in the signal peptide, W152C in the N-terminal domain (NTD), and L452R in the receptor-binding domain (RBD). Plasma from individuals vaccinated with a Wuhan-1 isolate-based messenger RNA vaccine or from convalescent individuals exhibited neutralizing titers that were reduced 2- to 3.5-fold against the B.1.427/B.1.429 variant relative to wild-type pseudoviruses. The L452R mutation reduced neutralizing activity in 14 of 34 RBD-specific monoclonal antibodies (mAbs). The S13I and W152C mutations resulted in total loss of neutralization for 10 of 10 NTD-specific mAbs because the NTD antigenic supersite was remodeled by a shift of the signal peptide cleavage site and the formation of a new disulfide bond, as revealed by mass spectrometry and structural studies.
Organizational Affiliation:
Department of Biochemistry, University of Washington, Seattle, WA 98195, USA.