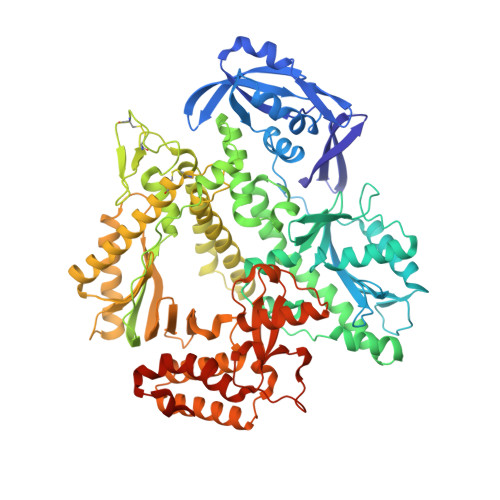
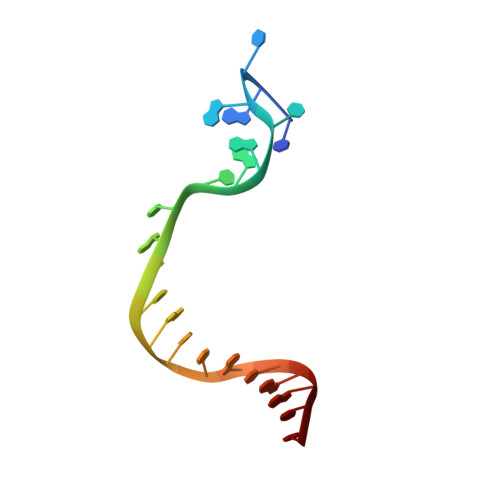
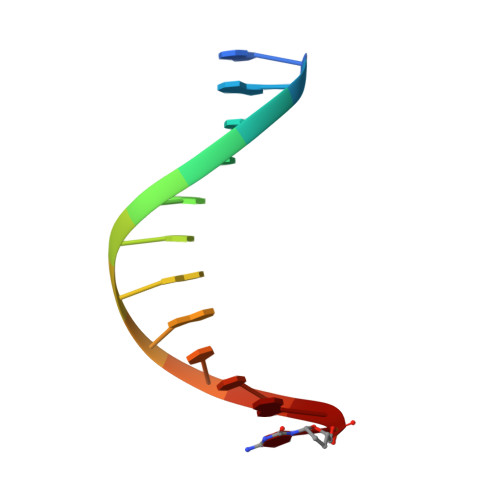
Structural Basis for The Recognition of Deaminated Nucleobases by An Archaeal DNA Polymerase.
Kropp, H.M., Ludmann, S., Diederichs, K., Betz, K., Marx, A.(2021) Chembiochem 22: 3060-3066
- PubMed: 34486208
- DOI: https://doi.org/10.1002/cbic.202100306
- Primary Citation of Related Structures:
7OM3, 7OMB, 7OMG - PubMed Abstract:
With increasing temperature, nucleobases in DNA become increasingly damaged by hydrolysis of exocyclic amines. The most prominent damage includes the conversion of cytosine to uracil and adenine to hypoxanthine. These damages are mutagenic and put the integrity of the genome at risk if not repaired appropriately. Several archaea live at elevated temperatures and thus, are exposed to a higher risk of deamination. Earlier studies have shown that DNA polymerases of archaea have the property of sensing deaminated nucleobases in the DNA template and thereby stalling the DNA synthesis during DNA replication providing another layer of DNA damage recognition and repair. However, the structural basis of uracil and hypoxanthine sensing by archaeal B-family DNA polymerases is sparse. Here we report on three new crystal structures of the archaeal B-family DNA polymerase from Thermococcus kodakarensis (KOD) DNA polymerase in complex with primer and template strands that have extended single stranded DNA template 5'-overhangs. These overhangs contain either the canonical nucleobases as well as uracil or hypoxanthine, respectively, and provide unprecedented structural insights into their recognition by archaeal B-family DNA polymerases.
Organizational Affiliation:
Department of Chemistry, University of Konstanz, Universitätsstraße 10, 78457, Konstanz, Germany.