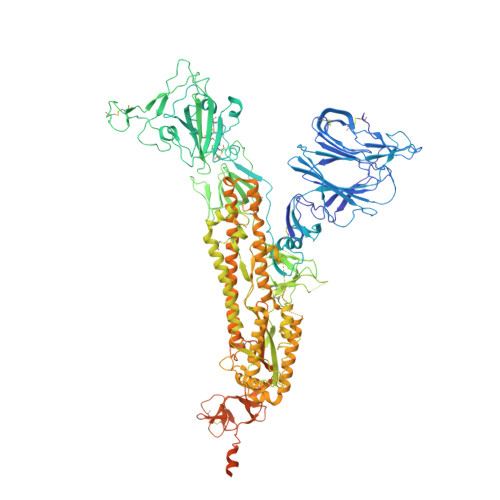
Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Walter, J.D., Scherer, M., Hutter, C.A.J., Garaeva, A.A., Zimmermann, I., Wyss, M., Rheinberger, J., Ruedin, Y., Earp, J.C., Egloff, P., Sorgenfrei, M., Hurlimann, L.M., Gonda, I., Meier, G., Remm, S., Thavarasah, S., van Geest, G., Bruggmann, R., Zimmer, G., Slotboom, D.J., Paulino, C., Plattet, P., Seeger, M.A.(2022) EMBO Rep 23: e54199-e54199
- PubMed: 35253970
- DOI: https://doi.org/10.15252/embr.202154199
- Primary Citation of Related Structures:
7P77, 7P78, 7P79, 7P7A, 7P7B - PubMed Abstract:
The ongoing COVID-19 pandemic represents an unprecedented global health crisis. Here, we report the identification of a synthetic nanobody (sybody) pair, Sb#15 and Sb#68, that can bind simultaneously to the SARS-CoV-2 spike RBD and efficiently neutralize pseudotyped and live viruses by interfering with ACE2 interaction. Cryo-EM confirms that Sb#15 and Sb#68 engage two spatially discrete epitopes, influencing rational design of bispecific and tri-bispecific fusion constructs that exhibit up to 100- and 1,000-fold increase in neutralization potency, respectively. Cryo-EM of the sybody-spike complex additionally reveals a novel up-out RBD conformation. While resistant viruses emerge rapidly in the presence of single binders, no escape variants are observed in the presence of the bispecific sybody. The multivalent bispecific constructs further increase the neutralization potency against globally circulating SARS-CoV-2 variants of concern. Our study illustrates the power of multivalency and biparatopic nanobody fusions for the potential development of therapeutic strategies that mitigate the emergence of new SARS-CoV-2 escape mutants.
Organizational Affiliation:
Institute of Medical Microbiology, University of Zurich, Zurich, Switzerland.