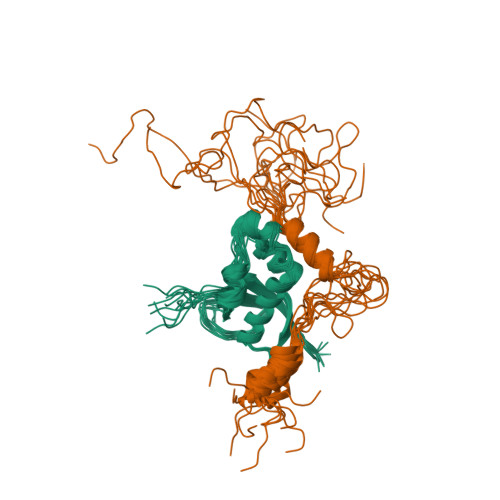
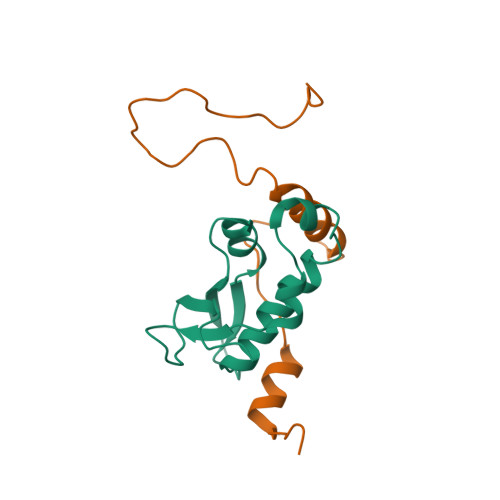
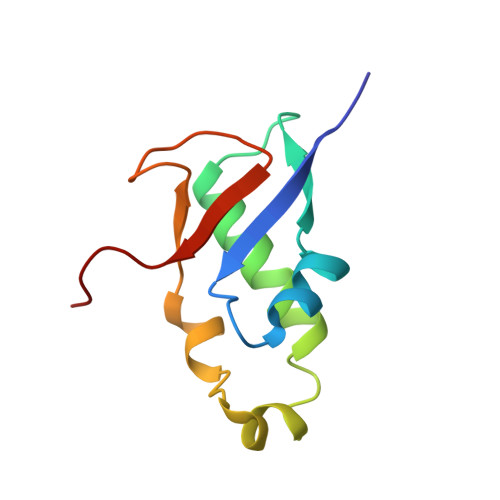
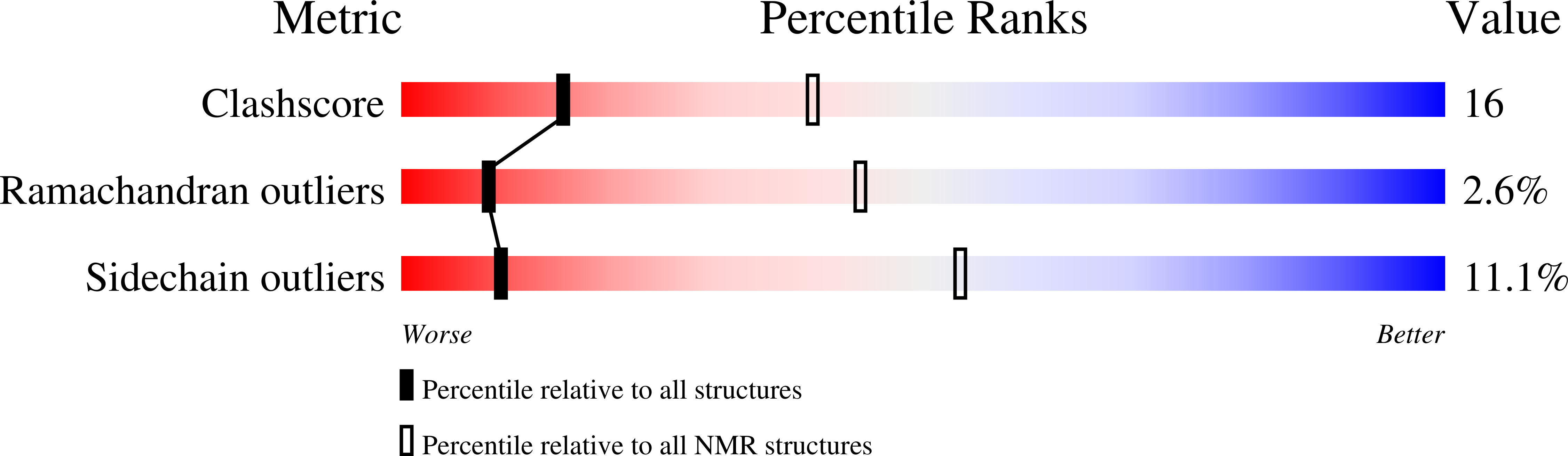The intrinsically disordered SARS-CoV-2 nucleoprotein in dynamic complex with its viral partner nsp3a.
Bessa, L.M., Guseva, S., Camacho-Zarco, A.R., Salvi, N., Maurin, D., Perez, L.M., Botova, M., Malki, A., Nanao, M., Jensen, M.R., Ruigrok, R.W.H., Blackledge, M.(2022) Sci Adv 8: eabm4034-eabm4034
- PubMed: 35044811
- DOI: https://doi.org/10.1126/sciadv.abm4034
- Primary Citation of Related Structures:
7PKU - PubMed Abstract:
The processes of genome replication and transcription of SARS-CoV-2 represent important targets for viral inhibition. Betacoronaviral nucleoprotein (N) is a highly dynamic cofactor of the replication-transcription complex (RTC), whose function depends on an essential interaction with the amino-terminal ubiquitin-like domain of nsp3 (Ubl1). Here, we describe this complex (dissociation constant - 30 to 200 nM) at atomic resolution. The interaction implicates two linear motifs in the intrinsically disordered linker domain (N3), a hydrophobic helix ( 219 LALLLLDRLNQL 230 ) and a disordered polar strand ( 243 GQTVTKKSAAEAS 255 ), that mutually engage to form a bipartite interaction, folding N3 around Ubl1. This results in substantial collapse in the dimensions of dimeric N, forming a highly compact molecular chaperone, that regulates binding to RNA, suggesting a key role of nsp3 in the association of N to the RTC. The identification of distinct linear motifs that mediate an important interaction between essential viral factors provides future targets for development of innovative strategies against COVID-19.
Organizational Affiliation:
Université Grenoble Alpes, CNRS, CEA, IBS, F-38000 Grenoble, France.