The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Liu, C., Zhou, D., Nutalai, R., Duyvesteyn, H.M.E., Tuekprakhon, A., Ginn, H.M., Dejnirattisai, W., Supasa, P., Mentzer, A.J., Wang, B., Case, J.B., Zhao, Y., Skelly, D.T., Chen, R.E., Johnson, S.A., Ritter, T.G., Mason, C., Malik, T., Temperton, N., Paterson, N.G., Williams, M.A., Hall, D.R., Clare, D.K., Howe, A., Goulder, P.J.R., Fry, E.E., Diamond, M.S., Mongkolsapaya, J., Ren, J., Stuart, D.I., Screaton, G.R.(2022) Cell Host Microbe 30: 53
- PubMed: 34921776
- DOI: https://doi.org/10.1016/j.chom.2021.11.013
- Primary Citation of Related Structures:
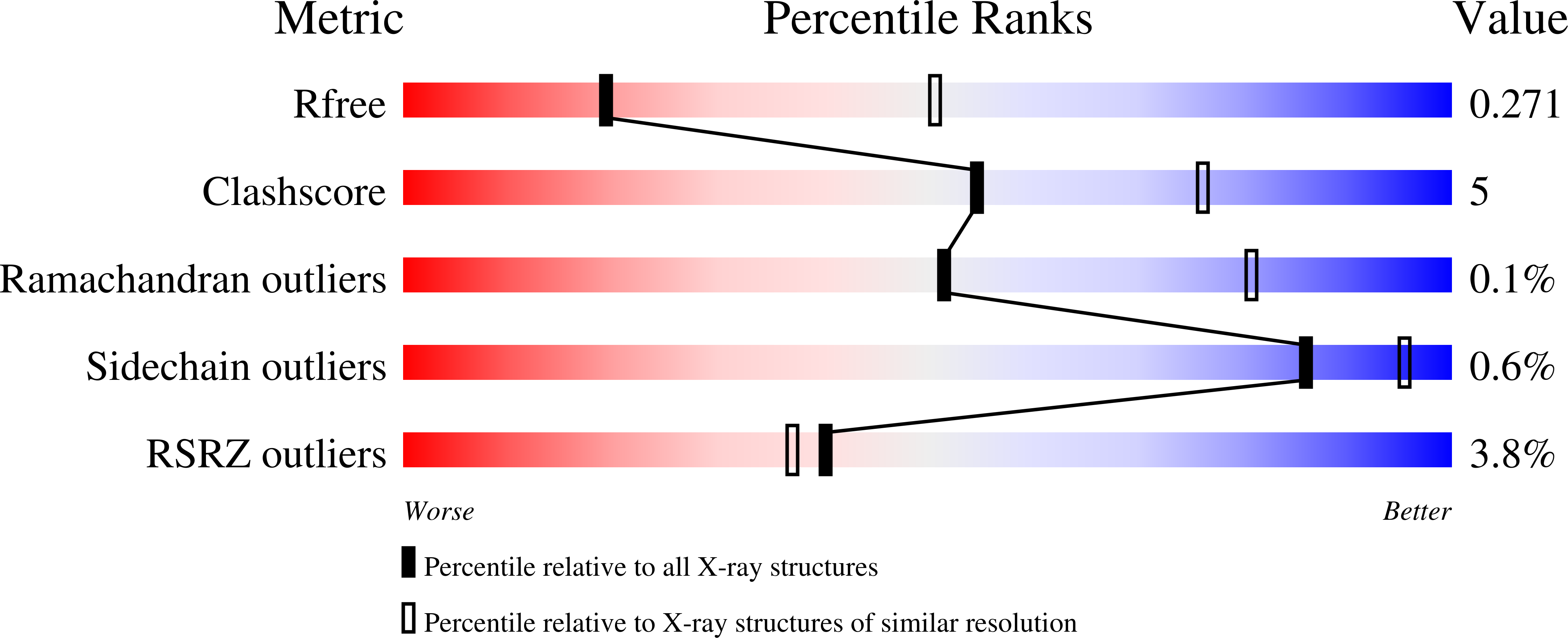
7PRY, 7PRZ, 7PS0, 7PS1, 7PS2, 7PS3, 7PS4, 7PS5, 7PS6, 7PS7, 7Q0G, 7Q0H, 7Q0I, 7Q6E, 7Q9F, 7Q9G, 7Q9I, 7Q9J, 7Q9K, 7Q9M, 7Q9P - PubMed Abstract:
Alpha-B.1.1.7, Beta-B.1.351, Gamma-P.1, and Delta-B.1.617.2 variants of SARS-CoV-2 express multiple mutations in the spike protein (S). These may alter the antigenic structure of S, causing escape from natural or vaccine-induced immunity. Beta is particularly difficult to neutralize using serum induced by early pandemic SARS-CoV-2 strains and is most antigenically separated from Delta. To understand this, we generated 674 mAbs from Beta-infected individuals and performed a detailed structure-function analysis of the 27 most potent mAbs: one binding the spike N-terminal domain (NTD), the rest the receptor-binding domain (RBD). Two of these RBD-binding mAbs recognize a neutralizing epitope conserved between SARS-CoV-1 and -2, while 18 target mutated residues in Beta: K417N, E484K, and N501Y. There is a major response to N501Y, including a public IgVH4-39 sequence, with E484K and K417N also targeted. Recognition of these key residues underscores why serum from Beta cases poorly neutralizes early pandemic and Delta viruses.
Organizational Affiliation:
Wellcome Centre for Human Genetics, Nuffield Department of Medicine, University of Oxford, Oxford, UK; Chinese Academy of Medical Science Oxford Institute, University of Oxford, Oxford, UK.