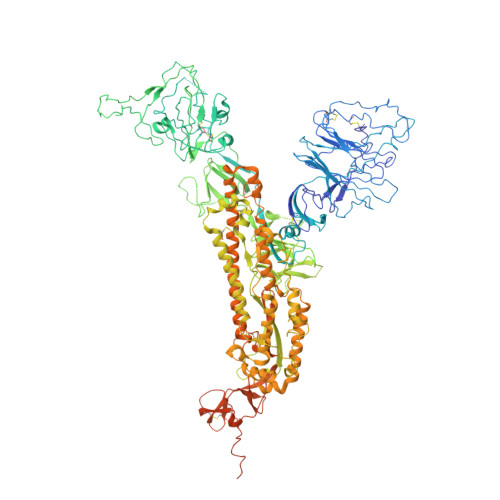
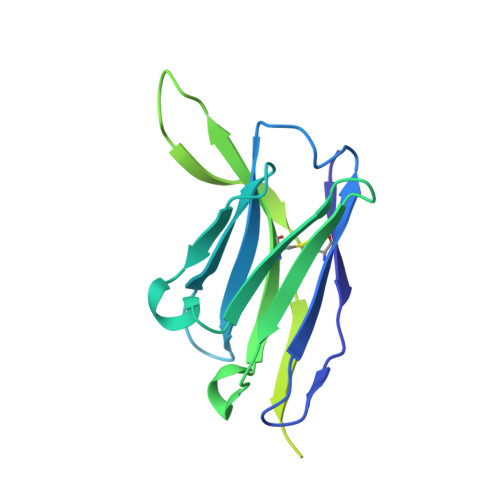
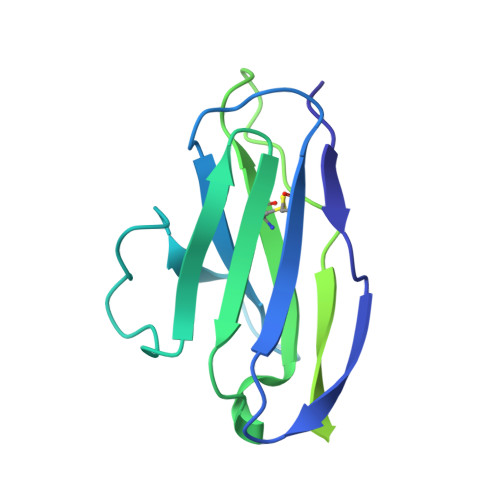
A Fc-enhanced NTD-binding non-neutralizing antibody delays virus spread and synergizes with a nAb to protect mice from lethal SARS-CoV-2 infection.
Beaudoin-Bussieres, G., Chen, Y., Ullah, I., Prevost, J., Tolbert, W.D., Symmes, K., Ding, S., Benlarbi, M., Gong, S.Y., Tauzin, A., Gasser, R., Chatterjee, D., Vezina, D., Goyette, G., Richard, J., Zhou, F., Stamatatos, L., McGuire, A.T., Charest, H., Roger, M., Pozharski, E., Kumar, P., Mothes, W., Uchil, P.D., Pazgier, M., Finzi, A.(2022) Cell Rep 38: 110368-110368
- PubMed: 35123652
- DOI: https://doi.org/10.1016/j.celrep.2022.110368
- Primary Citation of Related Structures:
7RQ6 - PubMed Abstract:
Emerging evidence indicates that both neutralizing and Fc-mediated effector functions of antibodies contribute to protection against SARS-CoV-2. It is unclear whether Fc-effector functions alone can protect against SARS-CoV-2. Here, we isolated CV3-13, a non-neutralizing antibody, from a convalescent individual with potent Fc-mediated effector functions. The cryoelectron microscopy structure of CV3-13 in complex with the SARS-CoV-2 spike reveals that the antibody binds from a distinct angle of approach to an N-terminal domain (NTD) epitope that only partially overlaps with the NTD supersite recognized by neutralizing antibodies. CV3-13 does not alter the replication dynamics of SARS-CoV-2 in K18-hACE2 mice, but its Fc-enhanced version significantly delays virus spread, neuroinvasion, and death in prophylactic settings. Interestingly, the combination of Fc-enhanced non-neutralizing CV3-13 with Fc-compromised neutralizing CV3-25 completely protects mice from lethal SARS-CoV-2 infection. Altogether, our data demonstrate that efficient Fc-mediated effector functions can potently contribute to the in vivo efficacy of anti-SARS-CoV-2 antibodies.
Organizational Affiliation:
Centre de recherche du CHUM, Montreal, QC H2X 0A9, Canada; Département de Microbiologie, Infectiologie et Immunologie, Université de Montréal, Montreal, QC H2X 0A9, Canada.