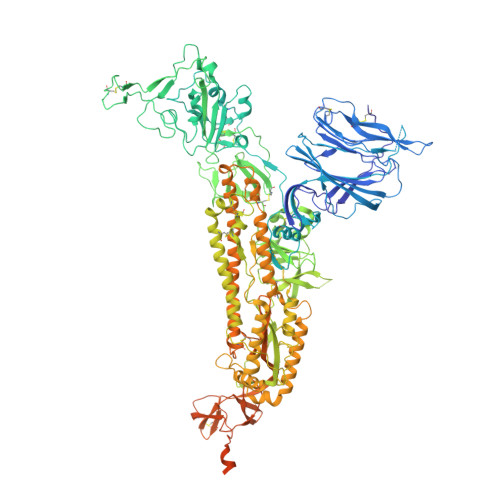
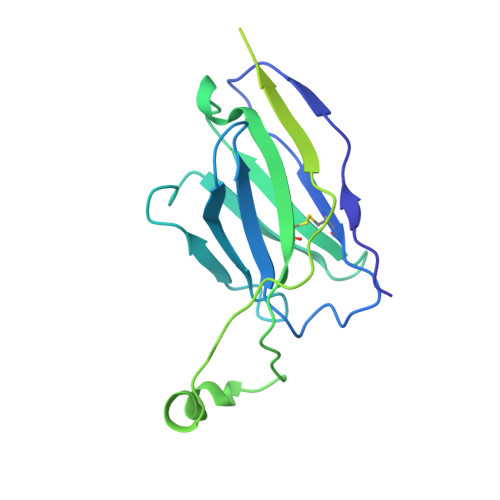
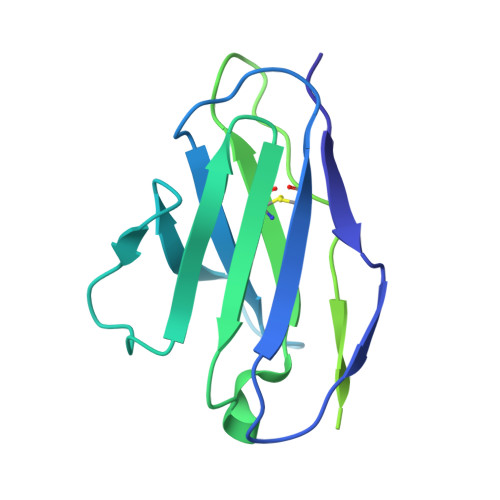
Neutralizing antibody 5-7 defines a distinct site of vulnerability in SARS-CoV-2 spike N-terminal domain.
Cerutti, G., Guo, Y., Wang, P., Nair, M.S., Wang, M., Huang, Y., Yu, J., Liu, L., Katsamba, P.S., Bahna, F., Reddem, E.R., Kwong, P.D., Ho, D.D., Sheng, Z., Shapiro, L.(2021) Cell Rep 37: 109928-109928
- PubMed: 34706271
- DOI: https://doi.org/10.1016/j.celrep.2021.109928
- Primary Citation of Related Structures:
7RW2 - PubMed Abstract:
Antibodies that potently neutralize SARS-CoV-2 target mainly the receptor-binding domain or the N-terminal domain (NTD). Over a dozen potently neutralizing NTD-directed antibodies have been studied structurally, and all target a single antigenic supersite in NTD (site 1). Here, we report the cryo-EM structure of a potent NTD-directed neutralizing antibody 5-7, which recognizes a site distinct from other potently neutralizing antibodies, inserting a binding loop into an exposed hydrophobic pocket between the two sheets of the NTD β sandwich. Interestingly, this pocket was previously identified as the binding site for hydrophobic molecules, including heme metabolites, but we observe that their presence does not substantially impede 5-7 recognition. Mirroring its distinctive binding, antibody 5-7 retains neutralization potency with many variants of concern (VOCs). Overall, we reveal that a hydrophobic pocket in NTD proposed for immune evasion can be used by the immune system for recognition.
Organizational Affiliation:
Zuckerman Mind Brain Behavior Institute, Columbia University, New York, NY 10027, USA.