A Systematic Survey of Reversibly Covalent Dipeptidyl Inhibitors of the SARS-CoV-2 Main Protease.
Geng, Z.Z., Atla, S., Shaabani, N., Vulupala, V., Yang, K.S., Alugubelli, Y.R., Khatua, K., Chen, P.H., Xiao, J., Blankenship, L.R., Ma, X.R., Vatansever, E.C., Cho, C.D., Ma, Y., Allen, R., Ji, H., Xu, S., Liu, W.R.(2023) J Med Chem 66: 11040-11055
- PubMed: 37561993
- DOI: https://doi.org/10.1021/acs.jmedchem.3c00221
- Primary Citation of Related Structures:
7SD9, 7SDA, 7SDC, 8STY, 8STZ - PubMed Abstract:
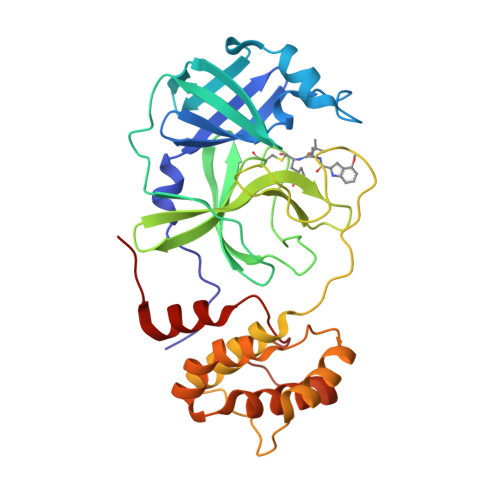
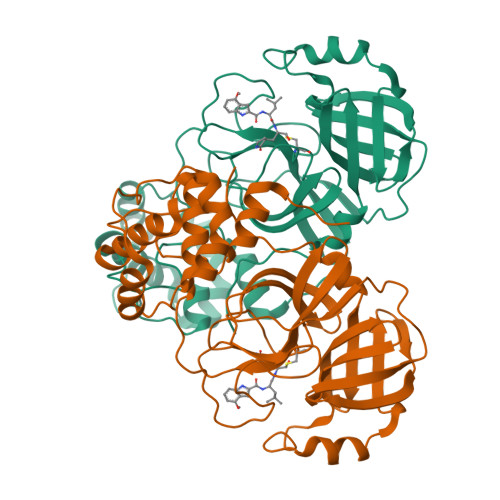
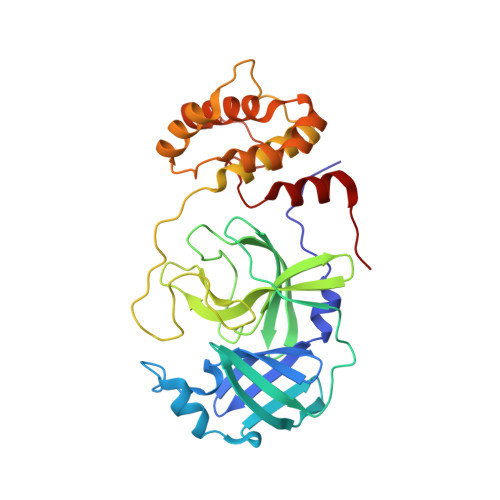
SARS-CoV-2, the COVID-19 pathogen, relies on its main protease (M Pro ) for replication and pathogenesis. M Pro is a demonstrated target for the development of antivirals for SARS-CoV-2. Past studies have systematically explored tripeptidyl inhibitors such as nirmatrelvir as M Pro inhibitors. However, dipeptidyl inhibitors especially those with a spiro residue at their P2 position have not been systematically investigated. In this work, we synthesized about 30 dipeptidyl M Pro inhibitors and characterized them on enzymatic inhibition potency, structures of their complexes with M Pro , cellular M Pro inhibition potency, antiviral potency, cytotoxicity, and in vitro metabolic stability. Our results indicated that M Pro has a flexible S2 pocket to accommodate inhibitors with a large P2 residue and revealed that dipeptidyl inhibitors with a large P2 spiro residue such as ( S )-2-azaspiro [4,4]nonane-3-carboxylate and ( S )-2-azaspiro[4,5]decane-3-carboxylate have favorable characteristics. One compound, MPI60, containing a P2 ( S )-2-azaspiro[4,4]nonane-3-carboxylate displayed high antiviral potency, low cellular cytotoxicity, and high in vitro metabolic stability.
Organizational Affiliation:
Department of Chemistry, Texas A&M Drug Discovery Laboratory, Texas A&M University, College Station, Texas 77843, United States.