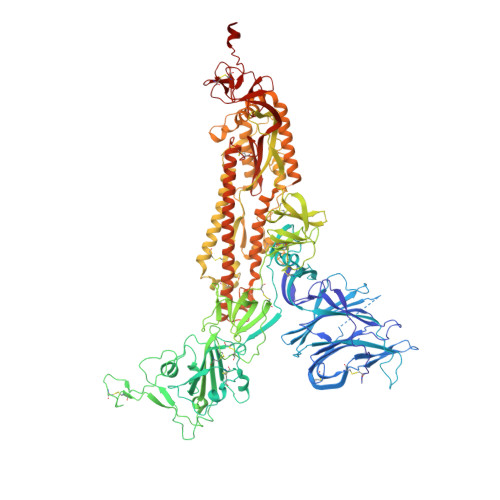
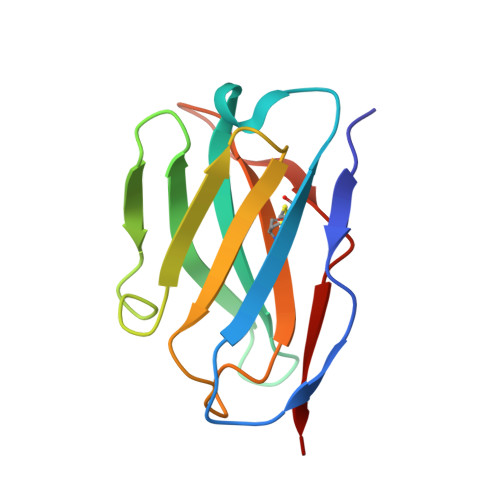
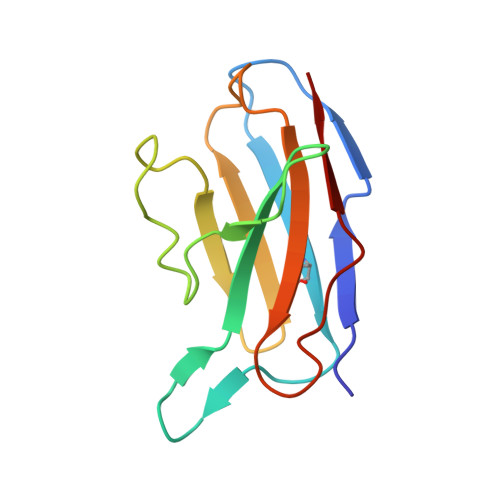
A protective antibody targeting the N-terminal domain of SARS-CoV-2 spike recognizes most emerging variants
Adams, L.J., VanBlargan, L.A., Liu, Z., Gilchuk, P., Zhao, H., Chen, R.E., Raju, S., Whitener, B., Shrihari, S., Jethva, P.N., Gross, M.L., Crowe, J.E., Whelan, S.P.J., Diamond, M.S., Fremont, D.H.To be published.