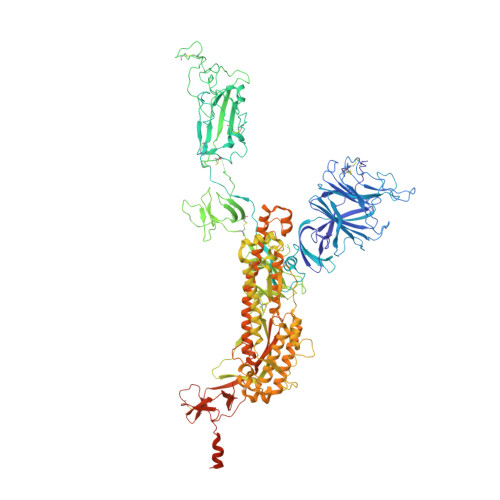
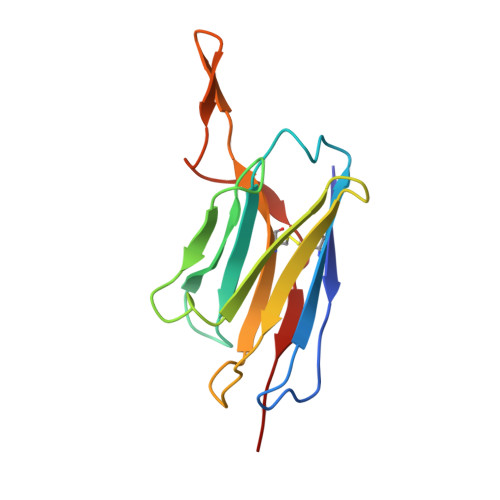
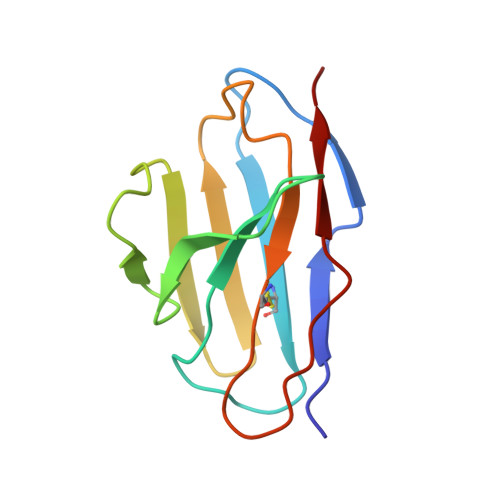
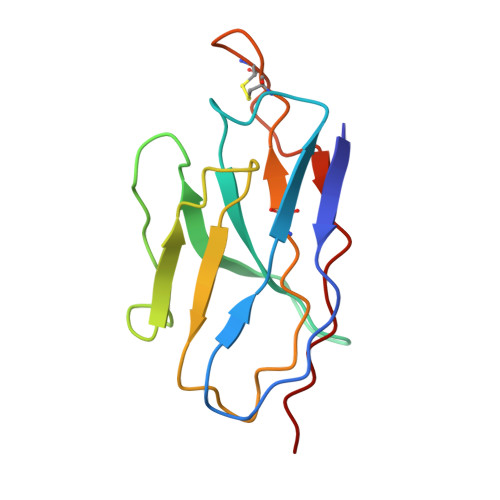
Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529.
Zhou, T., Wang, L., Misasi, J., Pegu, A., Zhang, Y., Harris, D.R., Olia, A.S., Talana, C.A., Yang, E.S., Chen, M., Choe, M., Shi, W., Teng, I.T., Creanga, A., Jenkins, C., Leung, K., Liu, T., Stancofski, E.D., Stephens, T., Zhang, B., Tsybovsky, Y., Graham, B.S., Mascola, J.R., Sullivan, N.J., Kwong, P.D.(2022) Science 376: eabn8897-eabn8897
- PubMed: 35324257
- DOI: https://doi.org/10.1126/science.abn8897
- Primary Citation of Related Structures:
7TB8, 7TBF, 7TC9, 7TCA, 7TCC, 7U0D - PubMed Abstract:
The rapid spread of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) B.1.1.529 (Omicron) variant and its resistance to neutralization by vaccinee and convalescent sera are driving a search for monoclonal antibodies with potent neutralization. To provide insight into effective neutralization, we determined cryo-electron microscopy structures and evaluated receptor binding domain (RBD) antibodies for their ability to bind and neutralize B.1.1.529. Mutations altered 16% of the B.1.1.529 RBD surface, clustered on an RBD ridge overlapping the angiotensin-converting enzyme 2 (ACE2)-binding surface and reduced binding of most antibodies. Substantial inhibitory activity was retained by select monoclonal antibodies-including A23-58.1, B1-182.1, COV2-2196, S2E12, A19-46.1, S309, and LY-CoV1404-that accommodated these changes and neutralized B.1.1.529. We identified combinations of antibodies with synergistic neutralization. The analysis revealed structural mechanisms for maintenance of potent neutralization against emerging variants.
Organizational Affiliation:
Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892, USA.