Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Kneller, D.W., Li, H., Phillips, G., Weiss, K.L., Zhang, Q., Arnould, M.A., Jonsson, C.B., Surendranathan, S., Parvathareddy, J., Blakeley, M.P., Coates, L., Louis, J.M., Bonnesen, P.V., Kovalevsky, A.(2022) Nat Commun 13: 2268
- PubMed: 35477935
- DOI: https://doi.org/10.1038/s41467-022-29915-z
- Primary Citation of Related Structures:
7SI9, 7TDU, 7TEH, 7TFR - PubMed Abstract:
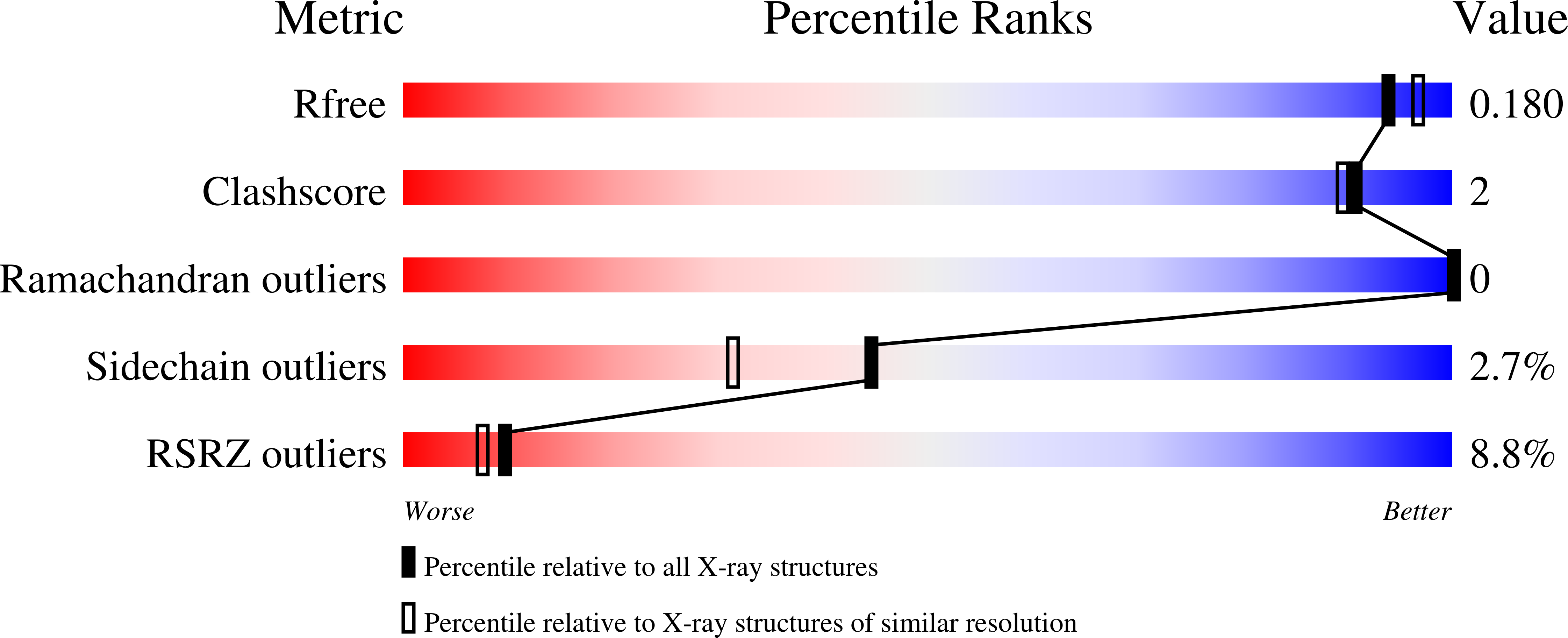
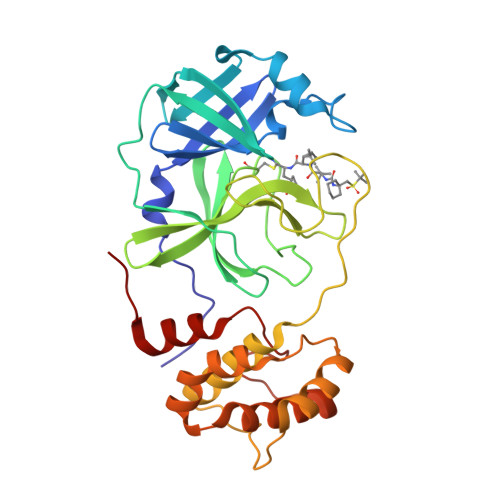
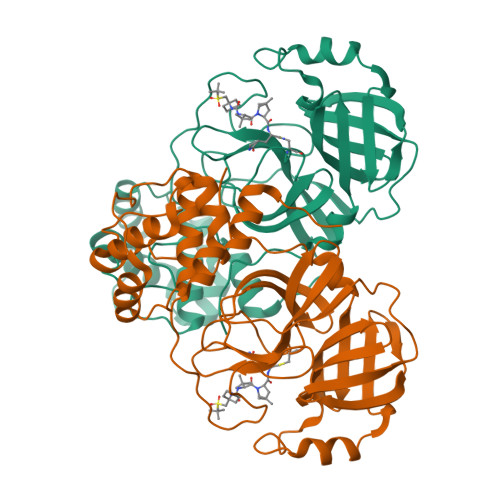
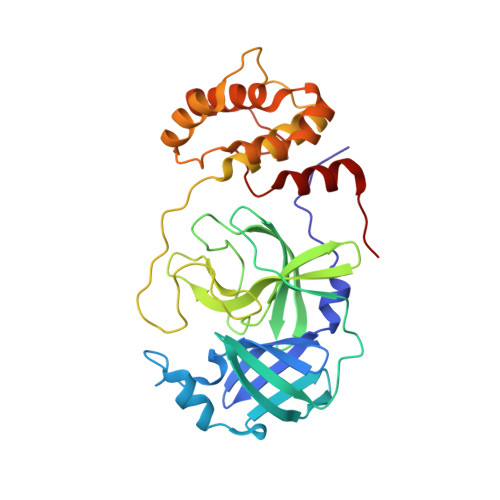
Emerging SARS-CoV-2 variants continue to threaten the effectiveness of COVID-19 vaccines, and small-molecule antivirals can provide an important therapeutic treatment option. The viral main protease (M pro ) is critical for virus replication and thus is considered an attractive drug target. We performed the design and characterization of three covalent hybrid inhibitors BBH-1, BBH-2 and NBH-2 created by splicing components of hepatitis C protease inhibitors boceprevir and narlaprevir, and known SARS-CoV-1 protease inhibitors. A joint X-ray/neutron structure of the M pro /BBH-1 complex demonstrates that a Cys145 thiolate reaction with the inhibitor's keto-warhead creates a negatively charged oxyanion. Protonation states of the ionizable residues in the M pro active site adapt to the inhibitor, which appears to be an intrinsic property of M pro . Structural comparisons of the hybrid inhibitors with PF-07321332 reveal unconventional F···O interactions of PF-07321332 with M pro which may explain its more favorable enthalpy of binding. BBH-1, BBH-2 and NBH-2 exhibit comparable antiviral properties in vitro relative to PF-07321332, making them good candidates for further design of improved antivirals.
Organizational Affiliation:
Neutron Scattering Division, Oak Ridge National Laboratory, Oak Ridge, TN, 37831, USA.