Controllable Phycobilin Modification: An Alternative Photoacclimation Response in Cryptophyte Algae.
Spangler, L.C., Yu, M., Jeffrey, P.D., Scholes, G.D.(2022) ACS Cent Sci 8: 340-350
- PubMed: 35350600
- DOI: https://doi.org/10.1021/acscentsci.1c01209
- Primary Citation of Related Structures:
7S96, 7S97, 7TJA, 7TLF - PubMed Abstract:
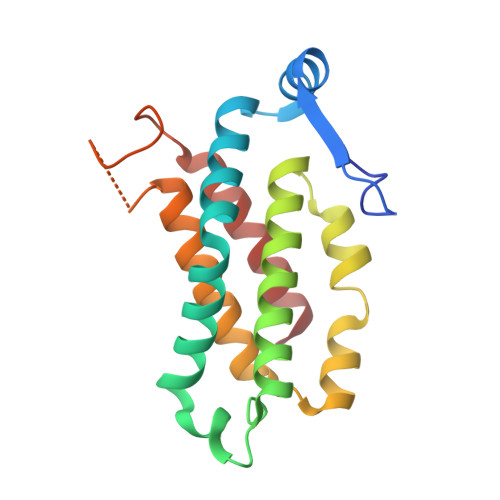
Cryptophyte algae are well-known for their ability to survive under low light conditions using their auxiliary light harvesting antennas, phycobiliproteins. Mainly acting to absorb light where chlorophyll cannot (500-650 nm), phycobiliproteins also play an instrumental role in helping cryptophyte algae respond to changes in light intensity through the process of photoacclimation. Until recently, photoacclimation in cryptophyte algae was only observed as a change in the cellular concentration of phycobiliproteins; however, an additional photoacclimation response was recently discovered that causes shifts in the phycobiliprotein absorbance peaks following growth under red, blue, or green light. Here, we reproduce this newly identified photoacclimation response in two species of cryptophyte algae and elucidate the origin of the response on the protein level. We compare isolated native and photoacclimated phycobiliproteins for these two species using spectroscopy and mass spectrometry, and we report the X-ray structures of each phycobiliprotein and the corresponding photoacclimated complex. We find that neither the protein sequences nor the protein structures are modified by photoacclimation. We conclude that cryptophyte algae change one chromophore in the phycobiliprotein β subunits in response to changes in the spectral quality of light. Ultrafast pump-probe spectroscopy shows that the energy transfer is weakly affected by photoacclimation.
Organizational Affiliation:
Department of Chemistry, Princeton University, Princeton, New Jersey 08544, United States.