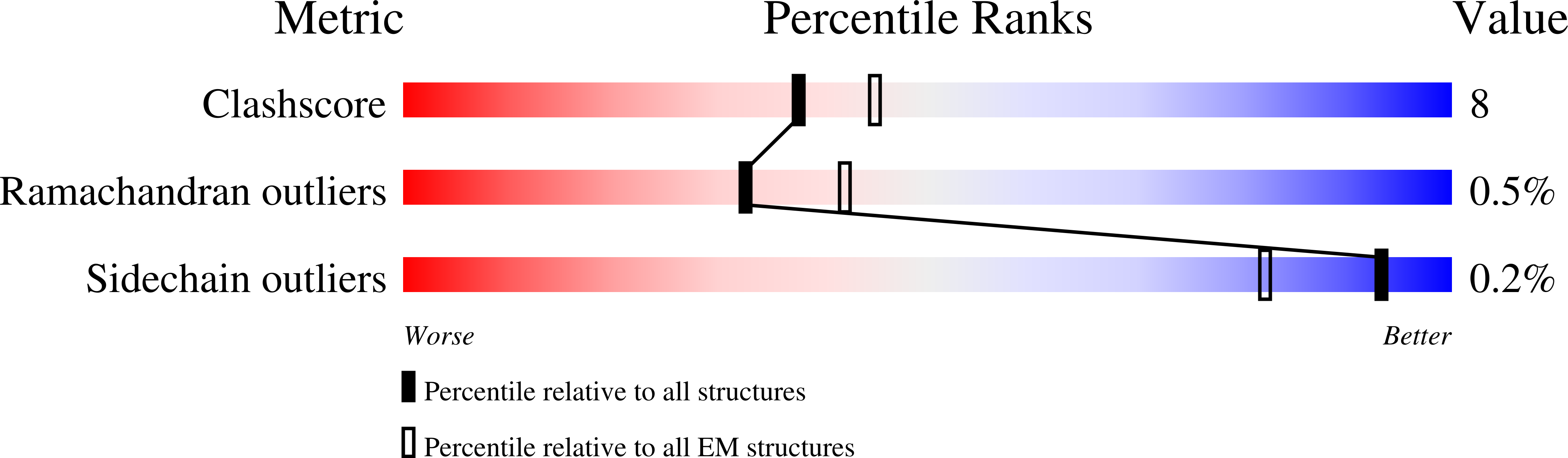Camel nanobodies broadly neutralize SARS-CoV-2 variants
Hong, J., Kwon, H.J., Cachau, R., Chen, C.Z., Dandey, V.P., Martin, N., Esposito, D., Ortega-Rodriguez, U., Xu, M., Borgnia, M.J., Xie, H., Ho, M.(2021) bioRxiv
- PubMed: 34751270
- DOI: https://doi.org/10.1101/2021.10.27.465996
- Primary Citation of Related Structures:
7TPR - PubMed Abstract:
With the emergence of SARS-CoV-2 variants, there is urgent need to develop broadly neutralizing antibodies. Here, we isolate two V H H nanobodies (7A3 and 8A2) from dromedary camels by phage display, which have high affinity for the receptor-binding domain (RBD) and broad neutralization activities against SARS-CoV-2 and its emerging variants. Cryo-EM complex structures reveal that 8A2 binds the RBD in its up mode and 7A3 inhibits receptor binding by uniquely targeting a highly conserved and deeply buried site in the spike regardless of the RBD conformational state. 7A3 at a dose of ≥5 mg/kg efficiently protects K18-hACE2 transgenic mice from the lethal challenge of B.1.351 or B.1.617.2, suggesting that the nanobody has promising therapeutic potentials to curb the COVID-19 surge with emerging SARS-CoV-2 variants. Dromedary camel ( Camelus dromedarius ) V H H phage libraries were built for isolation of the nanobodies that broadly neutralize SARS-CoV-2 variants.
Organizational Affiliation:
Laboratory of Molecular Biology, Center for Cancer Research, National Cancer Institute, National Institutes of Health; Bethesda, Maryland, 20891, USA.