Antibody targeting of conserved sites of vulnerability on the SARS-CoV-2 spike receptor-binding domain.
Sankhala, R.S., Dussupt, V., Chen, W.H., Bai, H., Martinez, E.J., Jensen, J.L., Rees, P.A., Hajduczki, A., Chang, W.C., Choe, M., Yan, L., Sterling, S.L., Swafford, I., Kuklis, C., Soman, S., King, J., Corbitt, C., Zemil, M., Peterson, C.E., Mendez-Rivera, L., Townsley, S.M., Donofrio, G.C., Lal, K.G., Tran, U., Green, E.C., Smith, C., de Val, N., Laing, E.D., Broder, C.C., Currier, J.R., Gromowski, G.D., Wieczorek, L., Rolland, M., Paquin-Proulx, D., van Dyk, D., Britton, Z., Rajan, S., Loo, Y.M., McTamney, P.M., Esser, M.T., Polonis, V.R., Michael, N.L., Krebs, S.J., Modjarrad, K., Joyce, M.G.(2024) Structure 32: 131-147.e7
- PubMed: 38157856
- DOI: https://doi.org/10.1016/j.str.2023.11.015
- Primary Citation of Related Structures:
7U8E, 8FAH, 8SGU, 8SMI, 8SMT - PubMed Abstract:
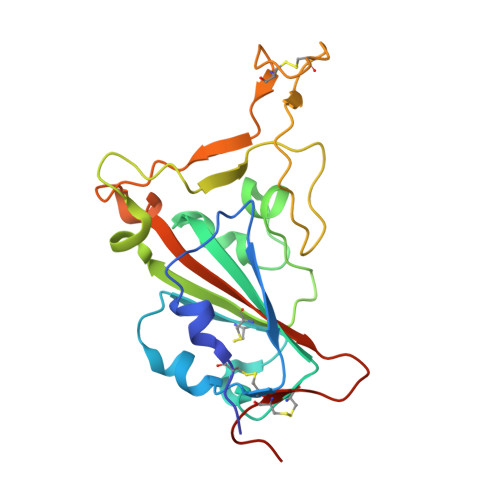
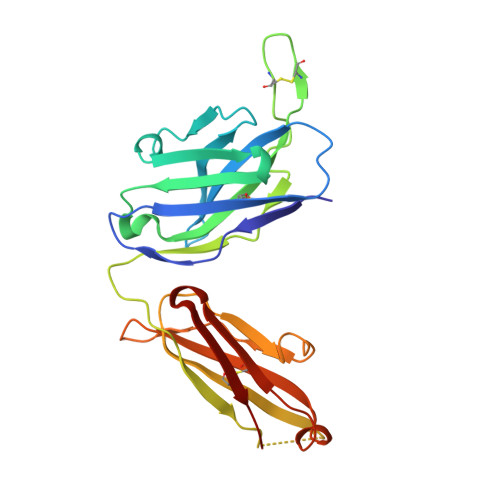
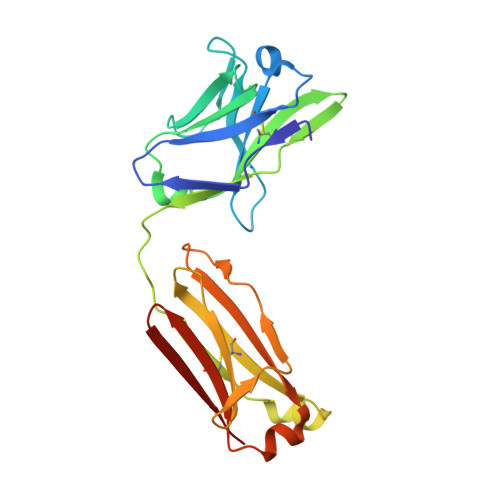
Given the continuous emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VoCs), immunotherapeutics that target conserved epitopes on the spike (S) glycoprotein have therapeutic advantages. Here, we report the crystal structure of the SARS-CoV-2 S receptor-binding domain (RBD) at 1.95 Å and describe flexibility and distinct conformations of the angiotensin-converting enzyme 2 (ACE2)-binding site. We identify a set of SARS-CoV-2-reactive monoclonal antibodies (mAbs) with broad RBD cross-reactivity including SARS-CoV-2 Omicron subvariants, SARS-CoV-1, and other sarbecoviruses and determine the crystal structures of mAb-RBD complexes with Ab246 and CR3022 mAbs targeting the class IV site, WRAIR-2134, which binds the recently designated class V epitope, and WRAIR-2123, the class I ACE2-binding site. The broad reactivity of class IV and V mAbs to conserved regions of SARS-CoV-2 VoCs and other sarbecovirus provides a framework for long-term immunotherapeutic development strategies.
Organizational Affiliation:
Emerging Infectious Diseases Branch, Walter Reed Army Institute of Research, Silver Spring, MD, USA; Henry M. Jackson Foundation for the Advancement of Military Medicine, Bethesda, MD, USA.