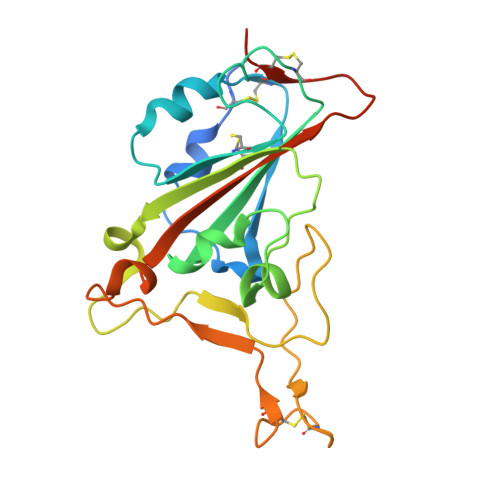
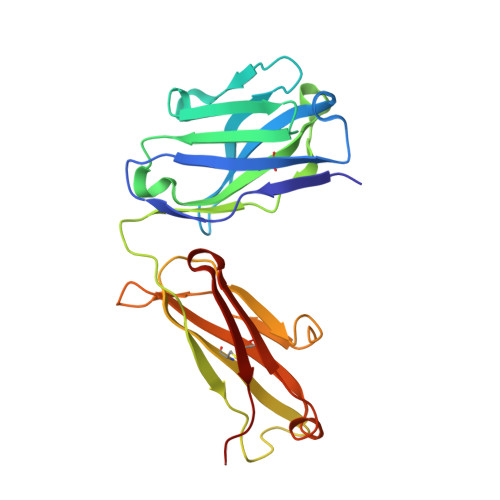
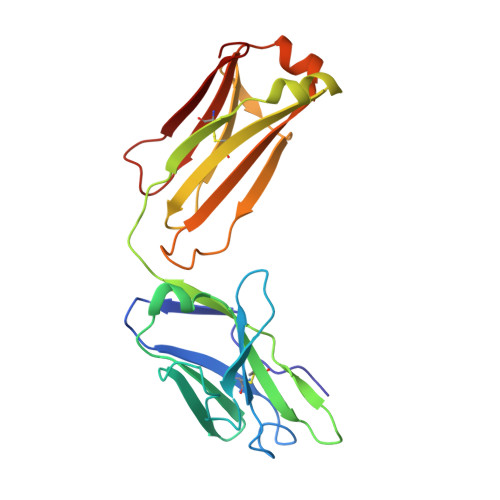
Molecular basis for antiviral activity of two pediatric neutralizing antibodies targeting SARS-CoV-2 Spike RBD.
Chen, Y., Prevost, J., Ullah, I., Romero, H., Lisi, V., Tolbert, W.D., Grover, J.R., Ding, S., Gong, S.Y., Beaudoin-Bussieres, G., Gasser, R., Benlarbi, M., Vezina, D., Anand, S.P., Chatterjee, D., Goyette, G., Grunst, M.W., Yang, Z., Bo, Y., Zhou, F., Beland, K., Bai, X., Zeher, A.R., Huang, R.K., Nguyen, D.N., Sherburn, R., Wu, D., Piszczek, G., Pare, B., Matthies, D., Xia, D., Richard, J., Kumar, P., Mothes, W., Cote, M., Uchil, P.D., Lavallee, V.P., Smith, M.A., Pazgier, M., Haddad, E., Finzi, A.(2023) iScience 26: 105783-105783
- PubMed: 36514310
- DOI: https://doi.org/10.1016/j.isci.2022.105783
- Primary Citation of Related Structures:
7UL0, 7UL1 - PubMed Abstract:
Neutralizing antibodies (NAbs) hold great promise for clinical interventions against SARS-CoV-2 variants of concern (VOCs). Understanding NAb epitope-dependent antiviral mechanisms is crucial for developing vaccines and therapeutics against VOCs. Here we characterized two potent NAbs, EH3 and EH8, isolated from an unvaccinated pediatric patient with exceptional plasma neutralization activity. EH3 and EH8 cross-neutralize the early VOCs and mediate strong Fc-dependent effector activity in vitro . Structural analyses of EH3 and EH8 in complex with the receptor-binding domain (RBD) revealed the molecular determinants of the epitope-driven protection and VOC evasion. While EH3 represents the prevalent IGHV3-53 NAb whose epitope substantially overlaps with the ACE2 binding site, EH8 recognizes a narrow epitope exposed in both RBD-up and RBD-down conformations. When tested in vivo, a single-dose prophylactic administration of EH3 fully protected stringent K18-hACE2 mice from lethal challenge with Delta VOC. Our study demonstrates that protective NAbs responses converge in pediatric and adult SARS-CoV-2 patients.
Organizational Affiliation:
Infectious Disease Division, Department of Medicine, Uniformed Services University of the Health Sciences, Bethesda, MD 20814-4712, USA.