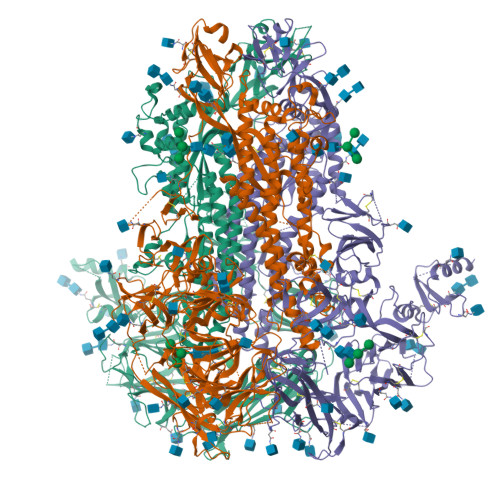
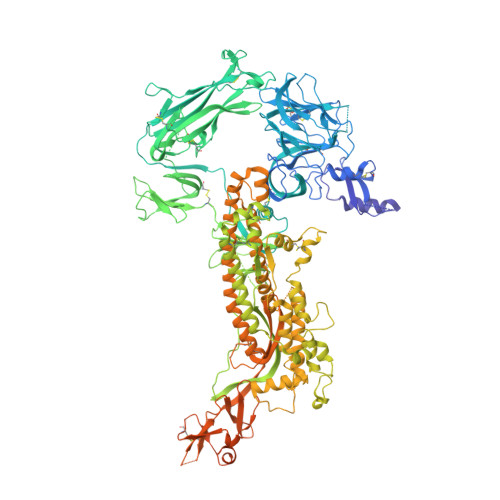
Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Tortorici, M.A., Walls, A.C., Joshi, A., Park, Y.J., Eguia, R.T., Miranda, M.C., Kepl, E., Dosey, A., Stevens-Ayers, T., Boeckh, M.J., Telenti, A., Lanzavecchia, A., King, N.P., Corti, D., Bloom, J.D., Veesler, D.(2022) Cell 185: 2279-2291.e17
- PubMed: 35700730
- DOI: https://doi.org/10.1016/j.cell.2022.05.019
- Primary Citation of Related Structures:
7U0L, 7US6, 7US9, 7USA, 7USB - PubMed Abstract:
The isolation of CCoV-HuPn-2018 from a child respiratory swab indicates that more coronaviruses are spilling over to humans than previously appreciated. We determined the structures of the CCoV-HuPn-2018 spike glycoprotein trimer in two distinct conformational states and showed that its domain 0 recognizes sialosides. We identified that the CCoV-HuPn-2018 spike binds canine, feline, and porcine aminopeptidase N (APN) orthologs, which serve as entry receptors, and determined the structure of the receptor-binding B domain in complex with canine APN. The introduction of an oligosaccharide at position N739 of human APN renders cells susceptible to CCoV-HuPn-2018 spike-mediated entry, suggesting that single-nucleotide polymorphisms might account for viral detection in some individuals. Human polyclonal plasma antibodies elicited by HCoV-229E infection and a porcine coronavirus monoclonal antibody inhibit CCoV-HuPn-2018 spike-mediated entry, underscoring the cross-neutralizing activity among ɑ-coronaviruses. These data pave the way for vaccine and therapeutic development targeting this zoonotic pathogen representing the eighth human-infecting coronavirus.
Organizational Affiliation:
Department of Biochemistry, University of Washington, Seattle, WA 98195, USA.