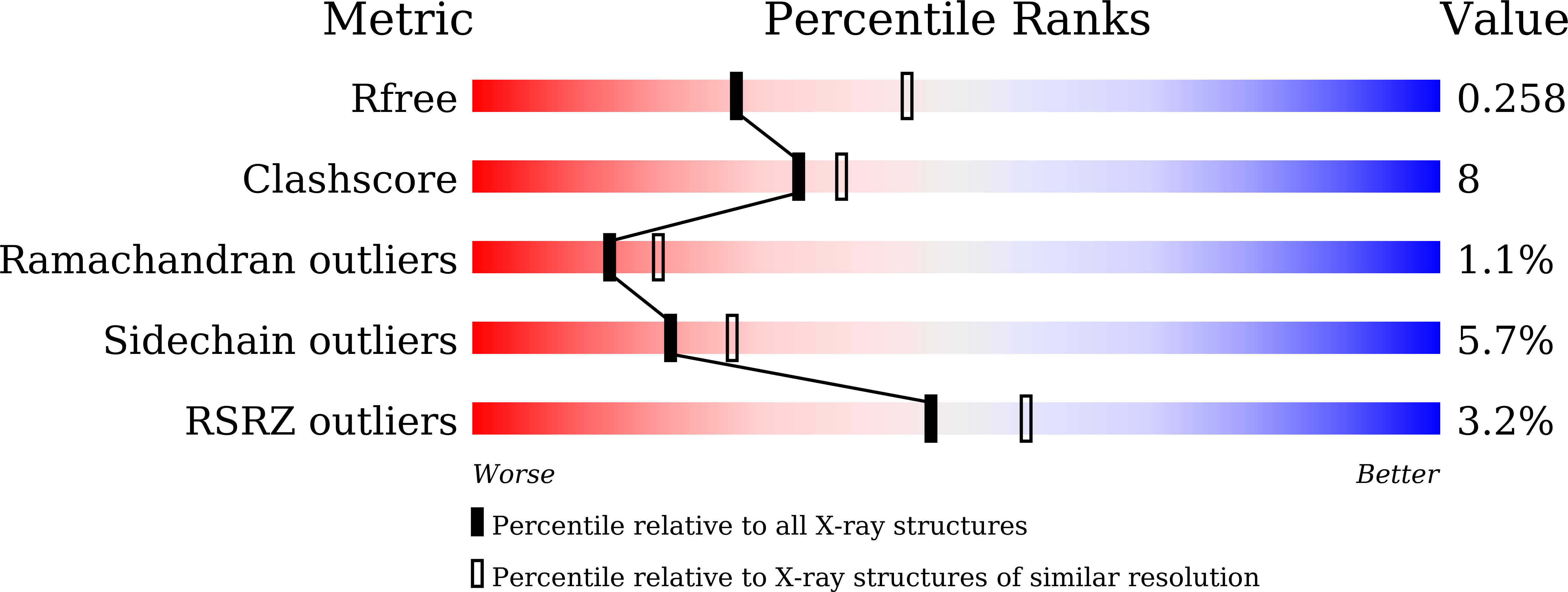
Regioselective stilbene O-methylations in Saccharinae grasses.
Lui, A.C.W., Pow, K.C., Lin, N., Lam, L.P.Y., Liu, G., Godwin, I.D., Fan, Z., Khoo, C.J., Tobimatsu, Y., Wang, L., Hao, Q., Lo, C.(2023) Nat Commun 14: 3462-3462
- PubMed: 37308495
- DOI: https://doi.org/10.1038/s41467-023-38908-5
- Primary Citation of Related Structures:
7VB8, 7WAQ, 7WAR, 7WAS - PubMed Abstract:
O-Methylated stilbenes are prominent nutraceuticals but rarely produced by crops. Here, the inherent ability of two Saccharinae grasses to produce regioselectively O-methylated stilbenes is reported. A stilbene O-methyltransferase, SbSOMT, is first shown to be indispensable for pathogen-inducible pterostilbene (3,5-bis-O-methylated) biosynthesis in sorghum (Sorghum bicolor). Phylogenetic analysis indicates the recruitment of genus-specific SOMTs from canonical caffeic acid O-methyltransferases (COMTs) after the divergence of Sorghum spp. from Saccharum spp. In recombinant enzyme assays, SbSOMT and COMTs regioselectively catalyze O-methylation of stilbene A-ring and B-ring respectively. Subsequently, SOMT-stilbene crystal structures are presented. Whilst SbSOMT shows global structural resemblance to SbCOMT, molecular characterizations illustrate two hydrophobic residues (Ile144/Phe337) crucial for substrate binding orientation leading to 3,5-bis-O-methylations in the A-ring. In contrast, the equivalent residues (Asn128/Asn323) in SbCOMT facilitate an opposite orientation that favors 3'-O-methylation in the B-ring. Consistently, a highly-conserved COMT is likely involved in isorhapontigenin (3'-O-methylated) formation in wounded wild sugarcane (Saccharum spontaneum). Altogether, our work reveals the potential of Saccharinae grasses as a source of O-methylated stilbenes, and rationalize the regioselectivity of SOMT activities for bioengineering of O-methylated stilbenes.
Organizational Affiliation:
School of Biological Sciences, The University of Hong Kong, Pokfulam, Hong Kong, China.