The Structure of the Porcine Deltacoronavirus Main Protease Reveals a Conserved Target for the Design of Antivirals.
Wang, F., Chen, C., Wang, Z., Han, X., Shi, P., Zhou, K., Liu, X., Xiao, Y., Cai, Y., Huang, J., Zhang, L., Yang, H.(2022) Viruses 14
- PubMed: 35336895
- DOI: https://doi.org/10.3390/v14030486
- Primary Citation of Related Structures:
7WKU - PubMed Abstract:
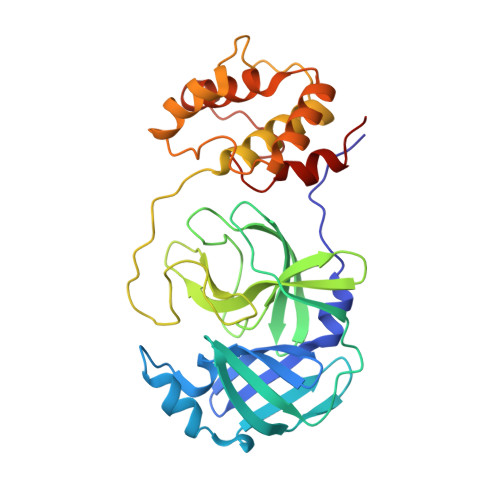
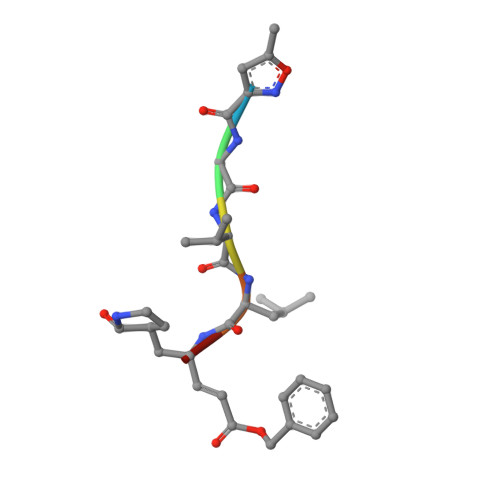
The existing zoonotic coronaviruses (CoVs) and viral genetic variants are important microbiological pathogens that cause severe disease in humans and animals. Currently, no effective broad-spectrum antiviral drugs against existing and emerging CoVs are available. The CoV main protease (M pro ) plays an essential role in viral replication, making it an ideal target for drug development. However, the structure of the Deltacoronavirus M pro is still unavailable. Porcine deltacoronavirus (PDCoV) is a novel CoV that belongs to the genus Deltacoronavirus and causes atrophic enteritis, severe diarrhea, vomiting and dehydration in pigs. Here, we determined the structure of PDCoV M pro complexed with a Michael acceptor inhibitor. Structural comparison showed that the backbone of PDCoV M pro is similar to those of alpha-, beta- and gamma-CoV M pro s. The substrate-binding pocket of M pro is well conserved in the subfamily Coronavirinae . In addition, we also observed that M pro s from the same genus adopted a similar conformation. Furthermore, the structure of PDCoV M pro in complex with a Michael acceptor inhibitor revealed the mechanism of its inhibition of PDCoV M pro . Our results provide a basis for the development of broad-spectrum antivirals against PDCoV and other CoVs.
Organizational Affiliation:
School of Life Sciences, Tianjin University, Tianjin 300072, China.