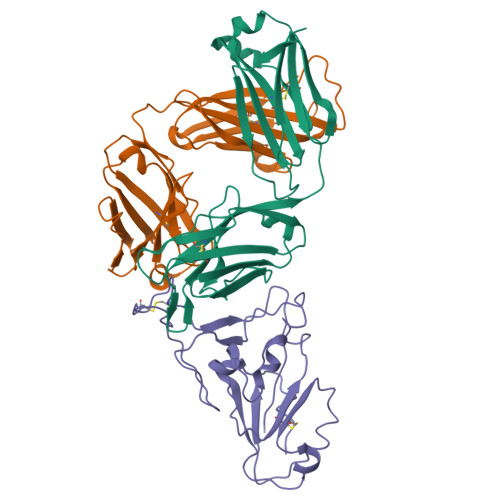
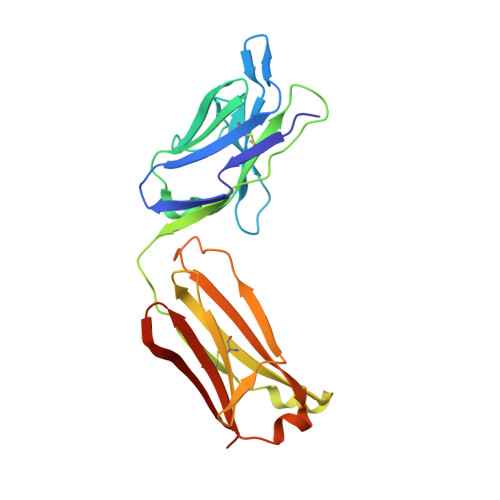
A broadly neutralizing monoclonal antibody overcomes the mutational landscape of emerging SARS-CoV-2 variants of concern.
Parray, H.A., Narayanan, N., Garg, S., Rizvi, Z.A., Shrivastava, T., Kushwaha, S., Singh, J., Murugavelu, P., Anantharaj, A., Mehdi, F., Raj, N., Singh, S., Dandotiya, J., Lukose, A., Jamwal, D., Kumar, S., Chiranjivi, A., Dhyani, S., Mishra, N., Kumar, S., Jakhar, K., Sonar, S., Panchal, A.K., Tripathy, M.R., Chowdhury, S.R., Ahmed, S., Samal, S., Mani, S., Bhattacharyya, S., Das, S., Sinha, S., Luthra, K., Batra, G., Sehgal, D., Medigeshi, G.R., Sharma, C., Awasthi, A., Garg, P.K., Nair, D.T., Kumar, R.(2022) PLoS Pathog 18: e1010994-e1010994
- PubMed: 36508467
- DOI: https://doi.org/10.1371/journal.ppat.1010994
- Primary Citation of Related Structures:
7WVL - PubMed Abstract:
The emergence of new variants of SARS-CoV-2 necessitates unremitting efforts to discover novel therapeutic monoclonal antibodies (mAbs). Here, we report an extremely potent mAb named P4A2 that can neutralize all the circulating variants of concern (VOCs) with high efficiency, including the highly transmissible Omicron. The crystal structure of the P4A2 Fab:RBD complex revealed that the residues of the RBD that interact with P4A2 are a part of the ACE2-receptor-binding motif and are not mutated in any of the VOCs. The pan coronavirus pseudotyped neutralization assay confirmed that the P4A2 mAb is specific for SARS-CoV-2 and its VOCs. Passive administration of P4A2 to K18-hACE2 transgenic mice conferred protection, both prophylactically and therapeutically, against challenge with VOCs. Overall, our data shows that, the P4A2 mAb has immense therapeutic potential to neutralize the current circulating VOCs. Due to the overlap between the P4A2 epitope and ACE2 binding site on spike-RBD, P4A2 may also be highly effective against a number of future variants.
Organizational Affiliation:
Translational Health Science and Technology Institute, NCR Biotech Science Cluster, Faridabad, Haryana, India.