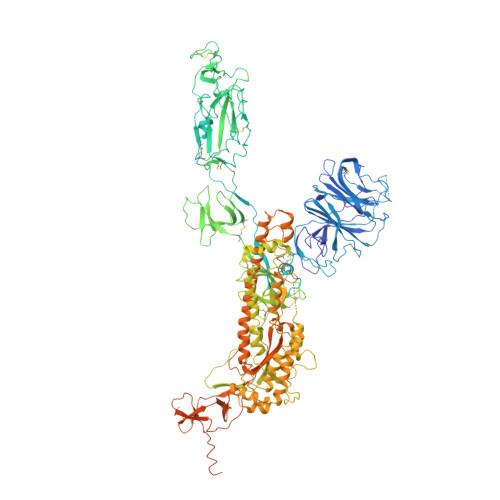
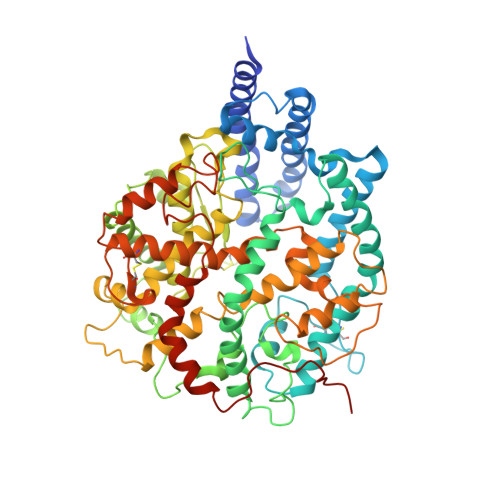
Molecular basis of receptor binding and antibody neutralization of Omicron.
Hong, Q., Han, W., Li, J., Xu, S., Wang, Y., Xu, C., Li, Z., Wang, Y., Zhang, C., Huang, Z., Cong, Y.(2022) Nature 604: 546-552
- PubMed: 35228716
- DOI: https://doi.org/10.1038/s41586-022-04581-9
- Primary Citation of Related Structures:
7WK2, 7WK8, 7WK9, 7WKA, 7WVN, 7WVO, 7WVP, 7WVQ - PubMed Abstract:
The SARS-CoV-2 Omicron variant exhibits striking immune evasion and is spreading rapidly worldwide. Understanding the structural basis of the high transmissibility and enhanced immune evasion of Omicron is of high importance. Here, using cryo-electron microscopy, we present both the closed and the open states of the Omicron spike (S) protein, which appear more compact than the counterparts of the G614 strain 1 , potentially related to enhanced inter-protomer and S1-S2 interactions induced by Omicron residue substitution. The closed state showing dominant population may indicate a conformational masking mechanism for the immune evasion of Omicron. Moreover, we captured three states for the Omicron S-ACE2 complex, revealing that the substitutions on the Omicron RBM result in new salt bridges and hydrogen bonds, more favourable electrostatic surface properties, and an overall strengthened S-ACE2 interaction, in line with the observed higher ACE2 affinity of Omicron S than of G614. Furthermore, we determined the structures of Omicron S in complex with the Fab of S3H3, an antibody that is able to cross-neutralize major variants of concern including Omicron, elucidating the structural basis for S3H3-mediated broad-spectrum neutralization. Our findings shed light on the receptor engagement and antibody neutralization or evasion of Omicron and may also inform the design of broadly effective vaccines against SARS-CoV-2.
Organizational Affiliation:
State Key Laboratory of Molecular Biology, National Center for Protein Science Shanghai, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, Shanghai, China.