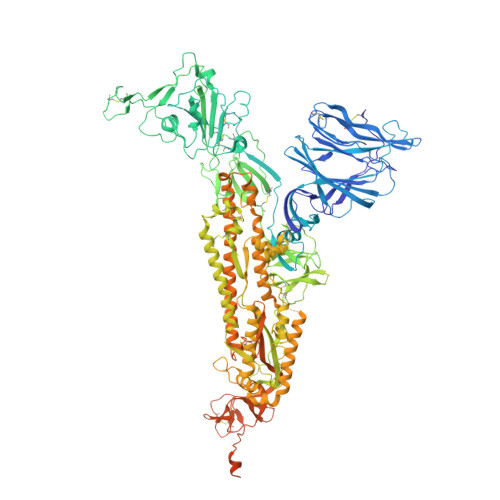
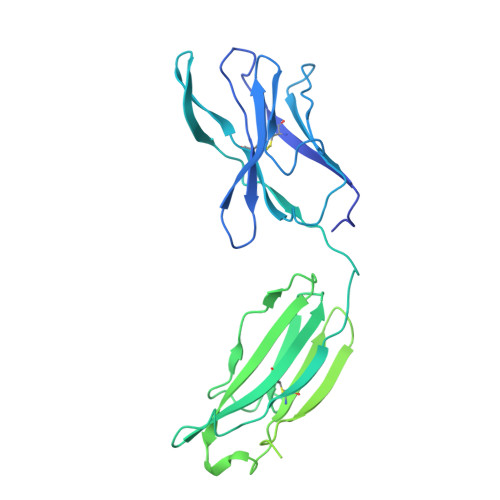
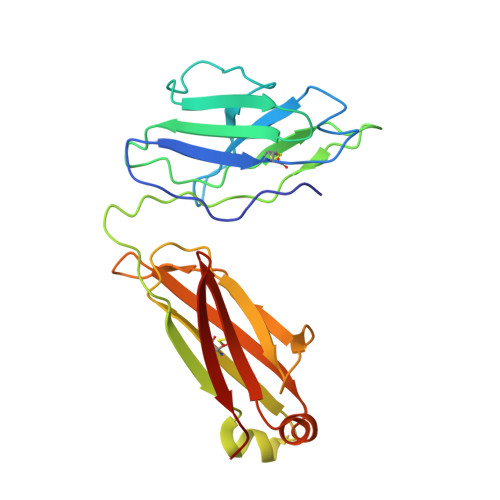
Broad ultra-potent neutralization of SARS-CoV-2 variants by monoclonal antibodies specific to the tip of RBD.
Ma, H., Guo, Y., Tang, H., Tseng, C.K., Wang, L., Zong, H., Wang, Z., He, Y., Chang, Y., Wang, S., Huang, H., Ke, Y., Yuan, Y., Wu, M., Zhang, Y., Drelich, A., Kempaiah, K.R., Peng, B.H., Wang, A., Yang, K., Yin, H., Liu, J., Yue, Y., Xu, W., Zhu, S., Ji, T., Zhang, X., Wang, Z., Li, G., Liu, G., Song, J., Mu, L., Xiang, Z., Song, Z., Chen, H., Bian, Y., Zhang, B., Chen, H., Zhang, J., Liao, Y., Zhang, L., Yang, L., Chen, Y., Gilly, J., Xiao, X., Han, L., Jiang, H., Xie, Y., Zhou, Q., Zhu, J.(2022) Cell Discov 8: 16-16
- PubMed: 35169121
- DOI: https://doi.org/10.1038/s41421-022-00381-7
- Primary Citation of Related Structures:
7X08 - PubMed Abstract:
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VOCs) continue to wreak havoc across the globe. Higher transmissibility and immunologic resistance of VOCs bring unprecedented challenges to epidemic extinguishment. Here we describe a monoclonal antibody, 2G1, that neutralizes all current VOCs and has surprising tolerance to mutations adjacent to or within its interaction epitope. Cryo-electron microscopy structure showed that 2G1 bound to the tip of receptor binding domain (RBD) of spike protein with small contact interface but strong hydrophobic effect, which resulted in nanomolar to sub-nanomolar affinities to spike proteins. The epitope of 2G1 on RBD partially overlaps with angiotensin converting enzyme 2 (ACE2) interface, which enables 2G1 to block interaction between RBD and ACE2. The narrow binding epitope but high affinity bestow outstanding therapeutic efficacy upon 2G1 that neutralized VOCs with sub-nanomolar half maximal inhibitory concentration in vitro. In SARS-CoV-2, Beta or Delta variant-challenged transgenic mice and rhesus macaque models, 2G1 protected animals from clinical illness and eliminated viral burden, without serious impact to animal safety. Mutagenesis experiments suggest that 2G1 is potentially capable of dealing with emerging SARS-CoV-2 variants in the future. This report characterized the therapeutic antibodies specific to the tip of spike against SARS-CoV-2 variants and highlights the potential clinical applications as well as for developing vaccine and cocktail therapy.
Organizational Affiliation:
Engineering Research Center of Cell and Therapeutic Antibody, Ministry of Education, China; School of Pharmacy, Shanghai Jiao Tong University, Shanghai, China.