Discovery of Chlorofluoroacetamide-Based Covalent Inhibitors for Severe Acute Respiratory Syndrome Coronavirus 2 3CL Protease.
Hirose, Y., Shindo, N., Mori, M., Onitsuka, S., Isogai, H., Hamada, R., Hiramoto, T., Ochi, J., Takahashi, D., Ueda, T., Caaveiro, J.M.M., Yoshida, Y., Ohdo, S., Matsunaga, N., Toba, S., Sasaki, M., Orba, Y., Sawa, H., Sato, A., Kawanishi, E., Ojida, A.(2022) J Med Chem 65: 13852-13865
- PubMed: 36229406
- DOI: https://doi.org/10.1021/acs.jmedchem.2c01081
- Primary Citation of Related Structures:
7XAR - PubMed Abstract:
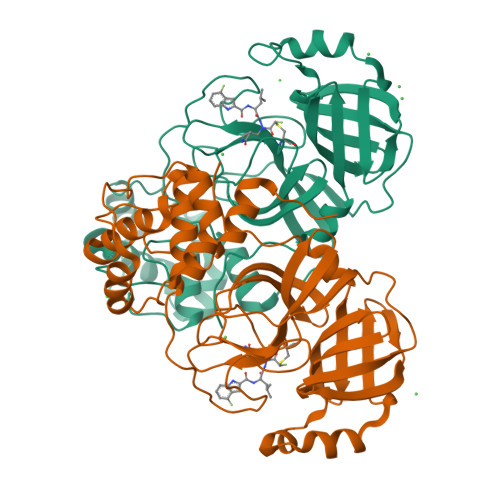
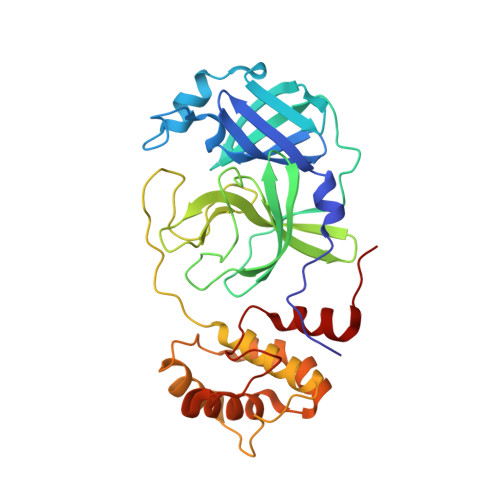
The coronavirus disease 2019 (COVID-19) pandemic has necessitated the development of antiviral agents against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). 3C-like protease (3CL pro ) is a promising target for COVID-19 treatment. Here, we report a new class of covalent inhibitors of 3CL pro that possess chlorofluoroacetamide (CFA) as a cysteine-reactive warhead. Based on an aza-peptide scaffold, we synthesized a series of CFA derivatives in enantiopure form and evaluated their biochemical efficiency. The data revealed that 8a ( YH-6 ) with the R configuration at the CFA unit strongly blocks SARS-CoV-2 replication in infected cells, and its potency is comparable to that of nirmatrelvir. X-ray structural analysis showed that YH-6 formed a covalent bond with Cys145 at the catalytic center of 3CL pro . The strong antiviral activity and favorable pharmacokinetic properties of YH-6 suggest its potential as a lead compound for the treatment of COVID-19.
Organizational Affiliation:
Graduate School of Pharmaceutical Sciences, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka812-8582, Japan.