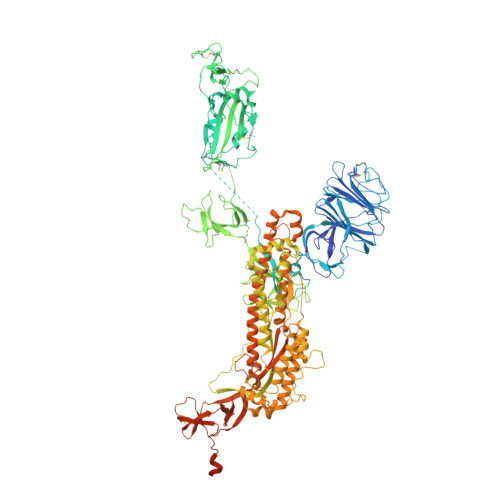
Structural basis of a two-antibody cocktail exhibiting highly potent and broadly neutralizing activities against SARS-CoV-2 variants including diverse Omicron sublineages.
Li, X., Pan, Y., Yin, Q., Wang, Z., Shan, S., Zhang, L., Yu, J., Qu, Y., Sun, L., Gui, F., Lu, J., Jing, Z., Wu, W., Huang, T., Shi, X., Li, J., Li, X., Li, D., Wang, S., Yang, M., Zhang, L., Duan, K., Liang, M., Yang, X., Wang, X.(2022) Cell Discov 8: 87-87
- PubMed: 36075908
- DOI: https://doi.org/10.1038/s41421-022-00449-4
- Primary Citation of Related Structures:
7XMX, 7XMZ, 7XST - PubMed Abstract:
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VOCs), especially the latest Omicron, have exhibited severe antibody evasion. Broadly neutralizing antibodies with high potency against Omicron are urgently needed for understanding the working mechanisms and developing therapeutic agents. In this study, we characterized the previously reported F61, which was isolated from convalescent patients infected with prototype SARS-CoV-2, as a broadly neutralizing antibody against all VOCs including Omicron BA.1, BA.1.1, BA.2, BA.3 and BA.4 sublineages by utilizing antigen binding and cell infection assays. We also identified and characterized another broadly neutralizing antibody D2 with epitope distinct from that of F61. More importantly, we showed that a combination of F61 with D2 exhibited synergy in neutralization and protecting mice from SARS-CoV-2 Delta and Omicron BA.1 variants. Cryo-Electron Microscopy (Cryo-EM) structures of the spike-F61 and spike-D2 binary complexes revealed the distinct epitopes of F61 and D2 at atomic level and the structural basis for neutralization. Cryo-EM structure of the Omicron-spike-F61-D2 ternary complex provides further structural insights into the synergy between F61 and D2. These results collectively indicated F61 and F61-D2 cocktail as promising therapeutic antibodies for combating SARS-CoV-2 variants including diverse Omicron sublineages.
Organizational Affiliation:
The Ministry of Education Key Laboratory of Protein Science, Beijing Advanced Innovation Center for Structural Biology, Beijing Frontier Research Center for Biological Structure, School of Life Sciences, Tsinghua University, Beijing, China.