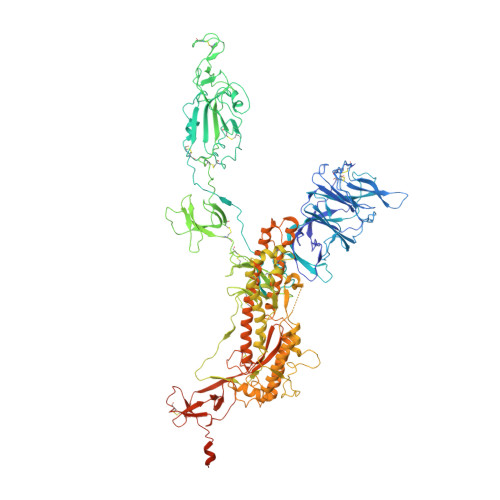
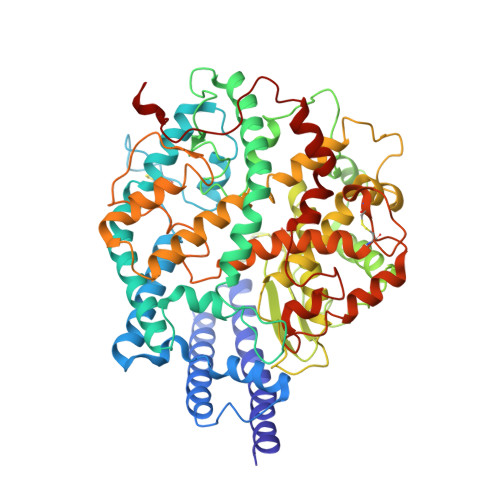
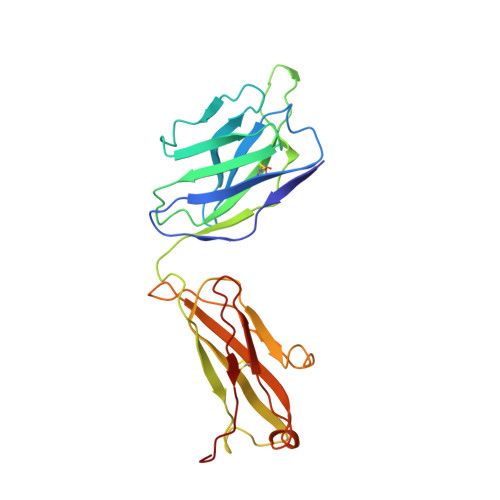
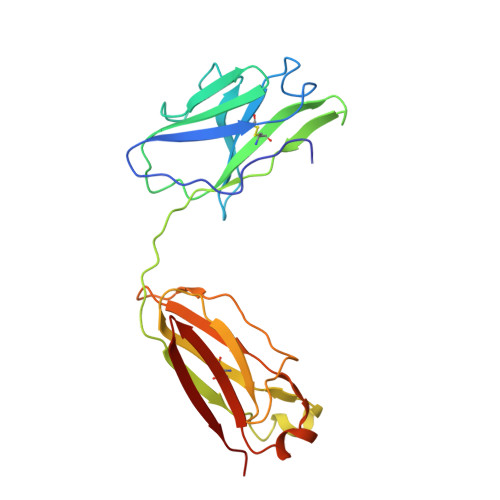
SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
He, P., Liu, B., Gao, X., Yan, Q., Pei, R., Sun, J., Chen, Q., Hou, R., Li, Z., Zhang, Y., Zhao, J., Sun, H., Feng, B., Wang, Q., Yi, H., Hu, P., Li, P., Zhang, Y., Chen, Z., Niu, X., Zhong, X., Jin, L., Liu, X., Qu, K., Ciazynska, K.A., Carter, A.P., Briggs, J.A.G., Chen, J., Liu, J., Chen, X., He, J., Chen, L., Xiong, X.(2022) Nat Microbiol 7: 1635-1649
- PubMed: 36151403
- DOI: https://doi.org/10.1038/s41564-022-01235-4
- Primary Citation of Related Structures:
7YDI, 7YDY, 7YE5, 7YE9, 7YEG - PubMed Abstract:
Population antibody response is thought to be important in selection of virus variants. We report that SARS-CoV-2 infection elicits a population immune response that is mediated by a lineage of VH1-69 germline antibodies. A representative antibody R1-32 from this lineage was isolated. By cryo-EM, we show that it targets a semi-cryptic epitope in the spike receptor-binding domain. Binding to this non-ACE2 competing epitope results in spike destruction, thereby inhibiting virus entry. On the basis of epitope location, neutralization mechanism and analysis of antibody binding to spike variants, we propose that recurrent substitutions at 452 and 490 are associated with immune evasion of the identified population antibody response. These substitutions, including L452R (present in the Delta variant), disrupt interactions mediated by the VH1-69-specific hydrophobic HCDR2 to impair antibody-antigen association, enabling variants to escape. The first Omicron variants were sensitive to antibody R1-32 but subvariants that harbour L452R quickly emerged and spread. Our results provide insights into how SARS-CoV-2 variants emerge and evade host immune responses.
Organizational Affiliation:
State Key Laboratory of Respiratory Disease, CAS Key Laboratory of Regenerative Biology, Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine, Guangdong Provincial Key Laboratory of Biocomputing, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou, China.