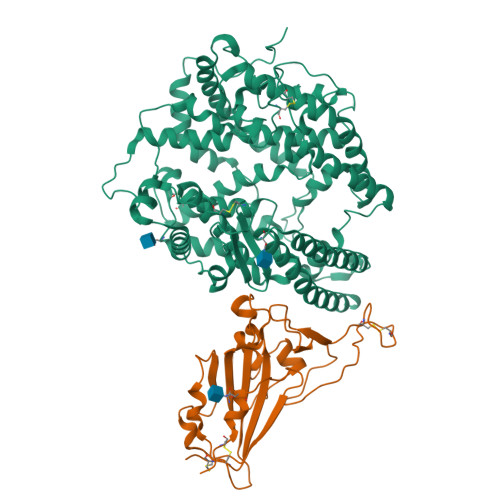
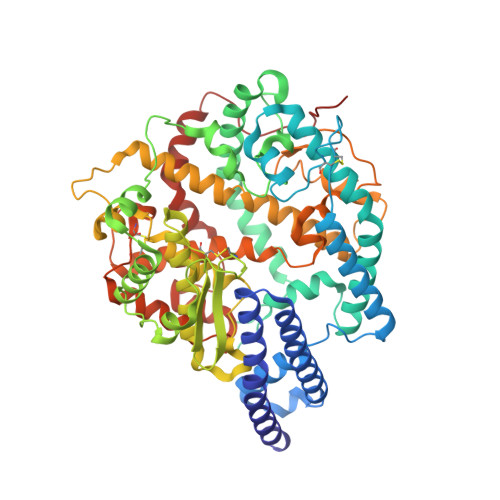
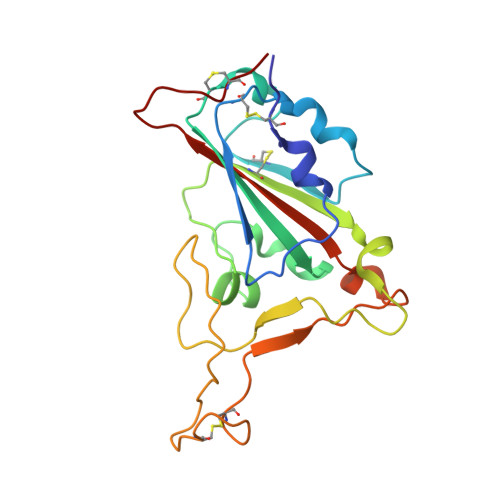
Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Zhao, Z., Xie, Y., Bai, B., Luo, C., Zhou, J., Li, W., Meng, Y., Li, L., Li, D., Li, X., Li, X., Wang, X., Sun, J., Xu, Z., Sun, Y., Zhang, W., Fan, Z., Zhao, X., Wu, L., Ma, J., Li, O.Y., Shang, G., Chai, Y., Liu, K., Wang, P., Gao, G.F., Qi, J.(2023) Nat Commun 14: 4405-4405
- PubMed: 37479708
- DOI: https://doi.org/10.1038/s41467-023-39942-z
- Primary Citation of Related Structures:
7YHW, 7YJ3, 7YV8, 7YVU, 8GRY, 8H06, 8H5C - PubMed Abstract:
Multiple SARS-CoV-2 Omicron sub-variants, such as BA.2, BA.2.12.1, BA.4, and BA.5, emerge one after another. BA.5 has become the dominant strain worldwide. Additionally, BA.2.75 is significantly increasing in some countries. Exploring their receptor binding and interspecies transmission risk is urgently needed. Herein, we examine the binding capacities of human and other 28 animal ACE2 orthologs covering nine orders towards S proteins of these sub-variants. The binding affinities between hACE2 and these sub-variants remain in the range as that of previous variants of concerns (VOCs) or interests (VOIs). Notably, R493Q reverse mutation enhances the bindings towards ACE2s from humans and many animals closely related to human life, suggesting an increased risk of cross-species transmission. Structures of S/hACE2 or RBD/hACE2 complexes for these sub-variants and BA.2 S binding to ACE2 of mouse, rat or golden hamster are determined to reveal the molecular basis for receptor binding and broader interspecies recognition.
Organizational Affiliation:
CAS Key Laboratory of Pathogen Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China.