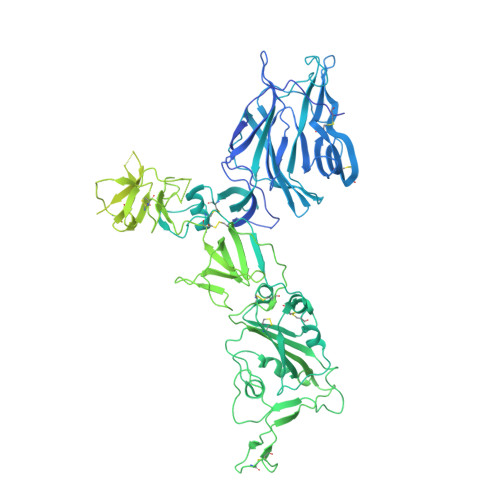
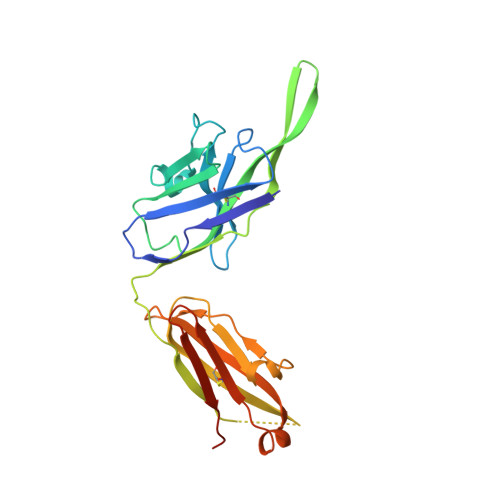
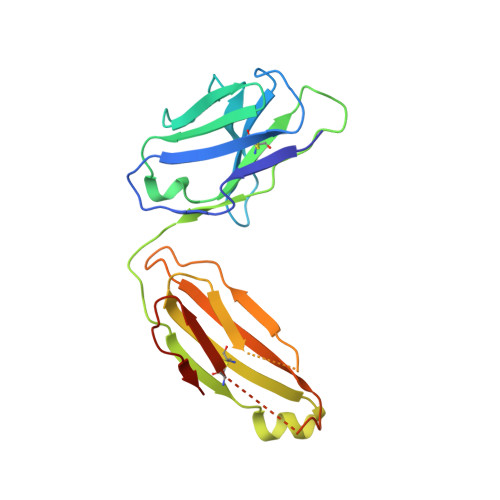
A neutralizing epitope on the SD1 domain of SARS-CoV-2 spike targeted following infection and vaccination.
Seow, J., Khan, H., Rosa, A., Calvaresi, V., Graham, C., Pickering, S., Pye, V.E., Cronin, N.B., Huettner, I., Malim, M.H., Politis, A., Cherepanov, P., Doores, K.J.(2022) Cell Rep 40: 111276-111276
- PubMed: 35981534
- DOI: https://doi.org/10.1016/j.celrep.2022.111276
- Primary Citation of Related Structures:
7ZBU - PubMed Abstract:
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike is the target for neutralizing antibodies elicited following both infection and vaccination. While extensive research has shown that the receptor binding domain (RBD) and, to a lesser extent, the N-terminal domain (NTD) are the predominant targets for neutralizing antibodies, identification of neutralizing epitopes beyond these regions is important for informing vaccine development and understanding antibody-mediated immune escape. Here, we identify a class of broadly neutralizing antibodies that bind an epitope on the spike subdomain 1 (SD1) and that have arisen from infection or vaccination. Using cryo-electron microscopy (cryo-EM) and hydrogen-deuterium exchange coupled to mass spectrometry (HDX-MS), we show that SD1-specific antibody P008_60 binds an epitope that is not accessible within the canonical prefusion states of the SARS-CoV-2 spike, suggesting a transient conformation of the viral glycoprotein that is vulnerable to neutralization.
Organizational Affiliation:
Department of Infectious Diseases, School of Immunology & Microbial Sciences, King's College London, London, UK.