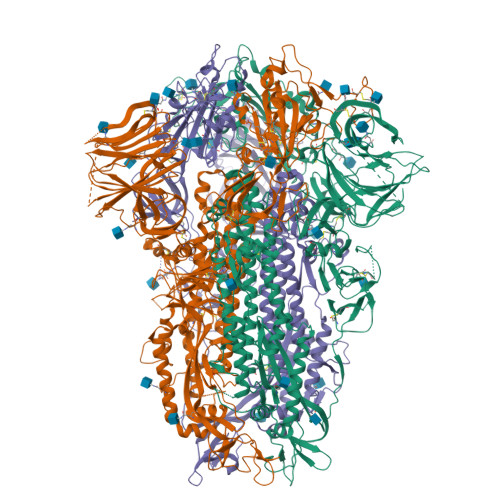
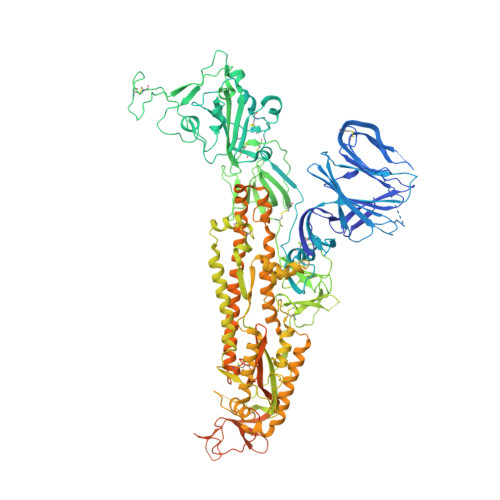
The free fatty acid-binding pocket is a conserved hallmark in pathogenic beta-coronavirus spike proteins from SARS-CoV to Omicron.
Toelzer, C., Gupta, K., Yadav, S.K.N., Hodgson, L., Williamson, M.K., Buzas, D., Borucu, U., Powers, K., Stenner, R., Vasileiou, K., Garzoni, F., Fitzgerald, D., Payre, C., Gautam, G., Lambeau, G., Davidson, A.D., Verkade, P., Frank, M., Berger, I., Schaffitzel, C.(2022) Sci Adv 8: eadc9179-eadc9179
- PubMed: 36417532
- DOI: https://doi.org/10.1126/sciadv.adc9179
- Primary Citation of Related Structures:
7ZH1, 7ZH2, 7ZH5 - PubMed Abstract:
As coronavirus disease 2019 (COVID-19) persists, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VOCs) emerge, accumulating spike (S) glycoprotein mutations. S receptor binding domain (RBD) comprises a free fatty acid (FFA)-binding pocket. FFA binding stabilizes a locked S conformation, interfering with virus infectivity. We provide evidence that the pocket is conserved in pathogenic β-coronaviruses (β-CoVs) infecting humans. SARS-CoV, MERS-CoV, SARS-CoV-2, and VOCs bind the essential FFA linoleic acid (LA), while binding is abolished by one mutation in common cold-causing HCoV-HKU1. In the SARS-CoV S structure, LA stabilizes the locked conformation, while the open, infectious conformation is devoid of LA. Electron tomography of SARS-CoV-2-infected cells reveals that LA treatment inhibits viral replication, resulting in fewer deformed virions. Our results establish FFA binding as a hallmark of pathogenic β-CoV infection and replication, setting the stage for FFA-based antiviral strategies to overcome COVID-19.
Organizational Affiliation:
School of Biochemistry, University of Bristol, 1 Tankard's Close, Bristol BS8 1TD, UK.