Cyanide Binding to [FeFe]-Hydrogenase Stabilizes the Alternative Configuration of the Proton Transfer Pathway.
Duan, J., Hemschemeier, A., Burr, D.J., Stripp, S.T., Hofmann, E., Happe, T.(2023) Angew Chem Int Ed Engl 62: e202216903-e202216903
- PubMed: 36464641
- DOI: https://doi.org/10.1002/anie.202216903
- Primary Citation of Related Structures:
8AIO, 8AJ6, 8ALN, 8AP2 - PubMed Abstract:
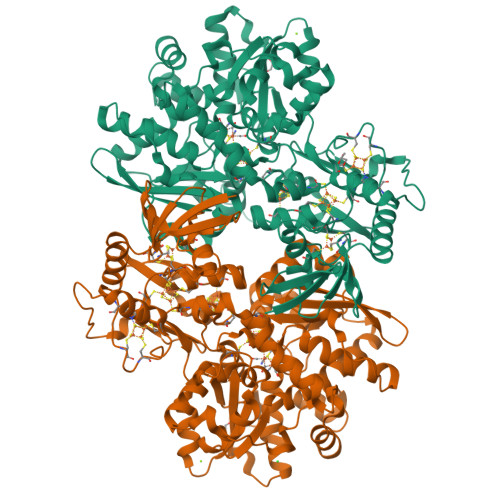
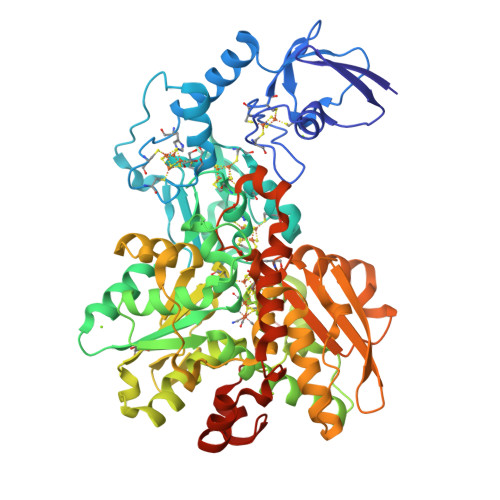
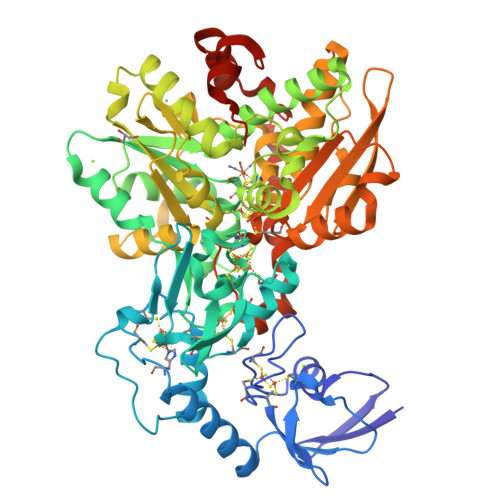
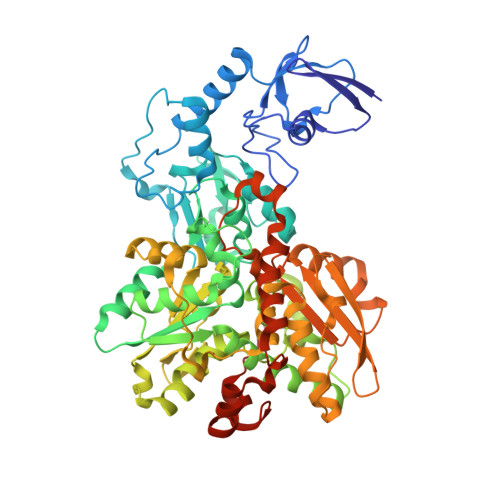
Hydrogenases are H 2 converting enzymes that harbor catalytic cofactors in which iron (Fe) ions are coordinated by biologically unusual carbon monoxide (CO) and cyanide (CN - ) ligands. Extrinsic CO and CN - , however, inhibit hydrogenases. The mechanism by which CN - binds to [FeFe]-hydrogenases is not known. Here, we obtained crystal structures of the CN - -treated [FeFe]-hydrogenase CpI from Clostridium pasteurianum. The high resolution of 1.39 Å allowed us to distinguish intrinsic CN - and CO ligands and to show that extrinsic CN - binds to the open coordination site of the cofactor where CO is known to bind. In contrast to other inhibitors, CN - treated crystals show conformational changes of conserved residues within the proton transfer pathway which could allow a direct proton transfer between E279 and S319. This configuration has been proposed to be vital for efficient proton transfer, but has never been observed structurally.
Organizational Affiliation:
Department of Plant Biochemistry, Faculty of Biology and Biotechnology, Photobiotechnology, Ruhr University Bochum, Universitätsstrasse 150, 44801, Bochum, Germany.