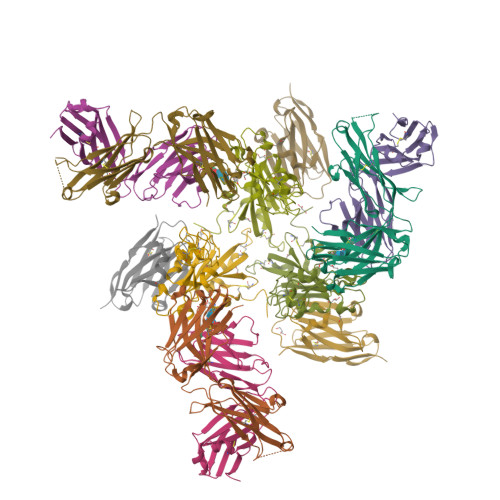
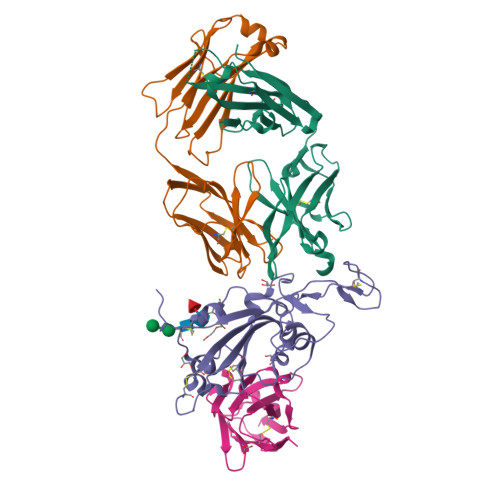
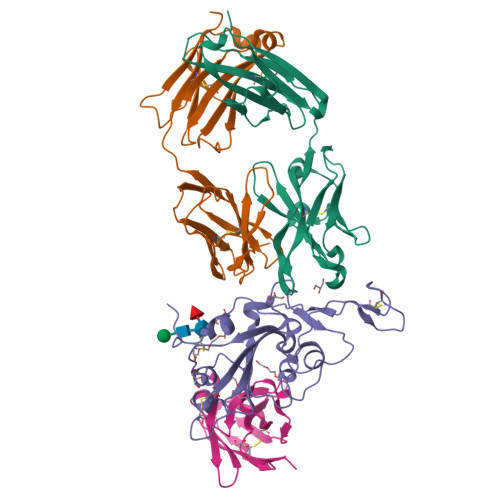
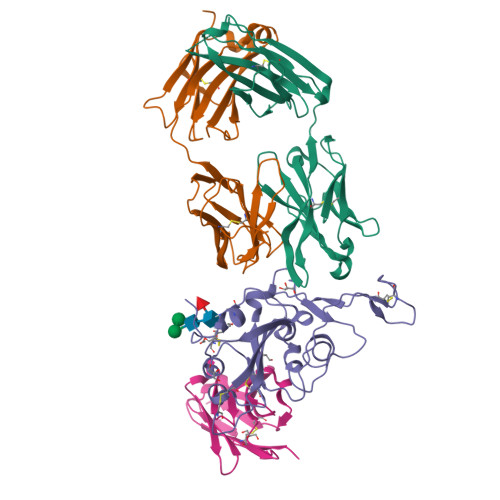
Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Dijokaite-Guraliuc, A., Das, R., Zhou, D., Ginn, H.M., Liu, C., Duyvesteyn, H.M.E., Huo, J., Nutalai, R., Supasa, P., Selvaraj, M., de Silva, T.I., Plowright, M., Newman, T.A.H., Hornsby, H., Mentzer, A.J., Skelly, D., Ritter, T.G., Temperton, N., Klenerman, P., Barnes, E., Dunachie, S.J., Roemer, C., Peacock, T.P., Paterson, N.G., Williams, M.A., Hall, D.R., Fry, E.E., Mongkolsapaya, J., Ren, J., Stuart, D.I., Screaton, G.R.(2023) Cell Rep 42: 112271-112271
- PubMed: 36995936
- DOI: https://doi.org/10.1016/j.celrep.2023.112271
- Primary Citation of Related Structures:
8BBN, 8BBO, 8BCZ, 8C3V - PubMed Abstract:
In November 2021, Omicron BA.1, containing a raft of new spike mutations, emerged and quickly spread globally. Intense selection pressure to escape the antibody response produced by vaccines or severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection then led to a rapid succession of Omicron sub-lineages with waves of BA.2 and then BA.4/5 infection. Recently, many variants have emerged such as BQ.1 and XBB, which carry up to 8 additional receptor-binding domain (RBD) amino acid substitutions compared with BA.2. We describe a panel of 25 potent monoclonal antibodies (mAbs) generated from vaccinees suffering BA.2 breakthrough infections. Epitope mapping shows potent mAb binding shifting to 3 clusters, 2 corresponding to early-pandemic binding hotspots. The RBD mutations in recent variants map close to these binding sites and knock out or severely knock down neutralization activity of all but 1 potent mAb. This recent mAb escape corresponds with large falls in neutralization titer of vaccine or BA.1, BA.2, or BA.4/5 immune serum.
Organizational Affiliation:
Wellcome Centre for Human Genetics, Nuffield Department of Medicine, University of Oxford, Oxford, UK.