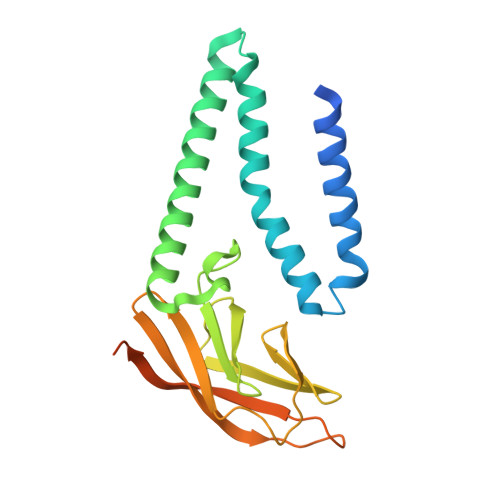
Structure of SARS-CoV-2 M protein in lipid nanodiscs.
Dolan, K.A., Dutta, M., Kern, D.M., Kotecha, A., Voth, G.A., Brohawn, S.G.(2022) Elife 11
- PubMed: 36264056
- DOI: https://doi.org/10.7554/eLife.81702
- Primary Citation of Related Structures:
8CTK - PubMed Abstract:
SARS-CoV-2 encodes four structural proteins incorporated into virions, spike (S), envelope (E), nucleocapsid (N), and membrane (M). M plays an essential role in viral assembly by organizing other structural proteins through physical interactions and directing them to sites of viral budding. As the most abundant protein in the viral envelope and a target of patient antibodies, M is a compelling target for vaccines and therapeutics. Still, the structure of M and molecular basis for its role in virion formation are unknown. Here, we present the cryo-EM structure of SARS-CoV-2 M in lipid nanodiscs to 3.5 Å resolution. M forms a 50 kDa homodimer that is structurally related to the SARS-CoV-2 ORF3a viroporin, suggesting a shared ancestral origin. Structural comparisons reveal how intersubunit gaps create a small, enclosed pocket in M and large open cavity in ORF3a, consistent with a structural role and ion channel activity, respectively. M displays a strikingly electropositive cytosolic surface that may be important for interactions with N, S, and viral RNA. Molecular dynamics simulations show a high degree of structural rigidity in a simple lipid bilayer and support a role for M homodimers in scaffolding viral assembly. Together, these results provide insight into roles for M in coronavirus assembly and structure.
Organizational Affiliation:
Biophysics Graduate Group, University of California, Berkeley, Berkeley, United States.