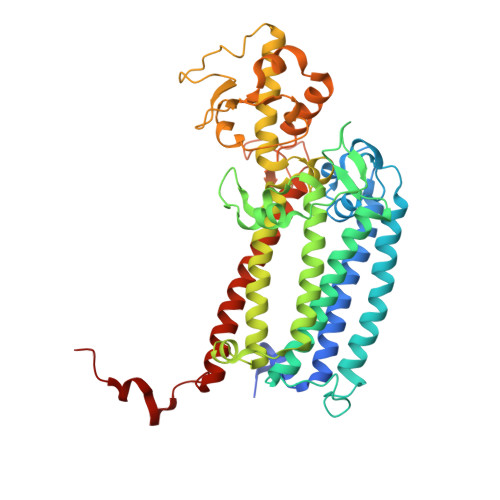
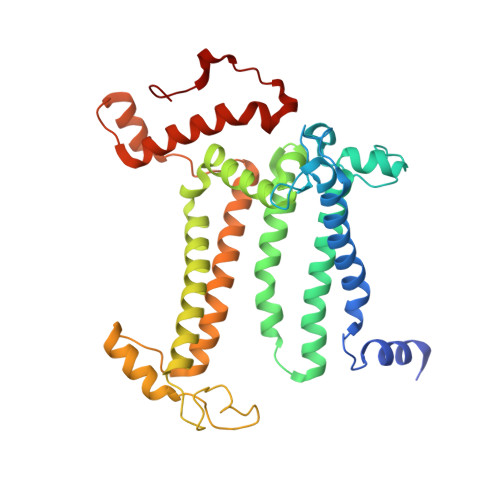
Structural evidence for intermediates during O 2 formation in photosystem II.
Bhowmick, A., Hussein, R., Bogacz, I., Simon, P.S., Ibrahim, M., Chatterjee, R., Doyle, M.D., Cheah, M.H., Fransson, T., Chernev, P., Kim, I.S., Makita, H., Dasgupta, M., Kaminsky, C.J., Zhang, M., Gatcke, J., Haupt, S., Nangca, I.I., Keable, S.M., Aydin, A.O., Tono, K., Owada, S., Gee, L.B., Fuller, F.D., Batyuk, A., Alonso-Mori, R., Holton, J.M., Paley, D.W., Moriarty, N.W., Mamedov, F., Adams, P.D., Brewster, A.S., Dobbek, H., Sauter, N.K., Bergmann, U., Zouni, A., Messinger, J., Kern, J., Yano, J., Yachandra, V.K.(2023) Nature 617: 629-636
- PubMed: 37138085
- DOI: https://doi.org/10.1038/s41586-023-06038-z
- Primary Citation of Related Structures:
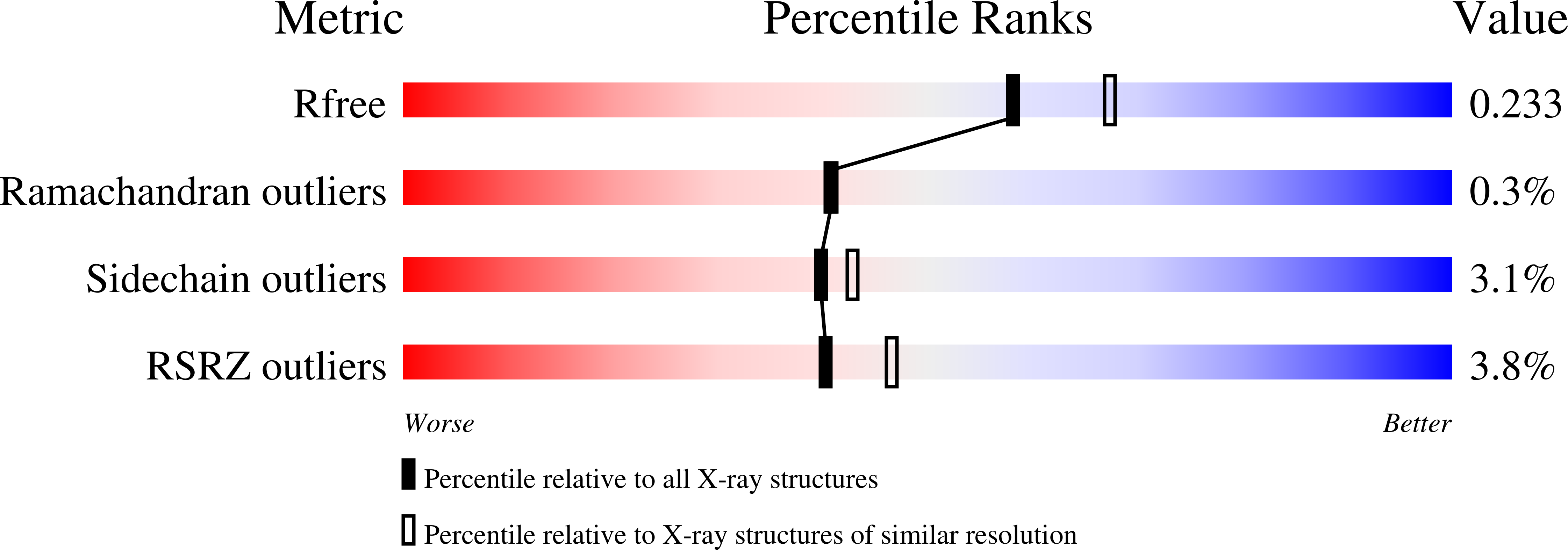
8EZ5, 8F4C, 8F4D, 8F4E, 8F4F, 8F4G, 8F4H, 8F4I, 8F4J, 8F4K - PubMed Abstract:
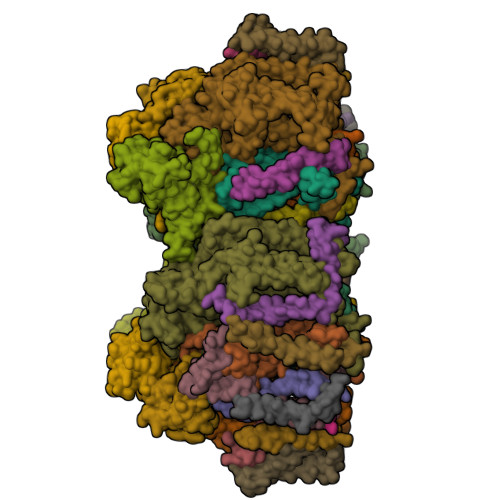
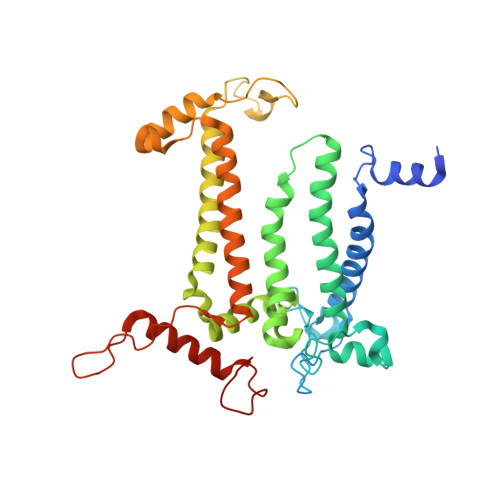
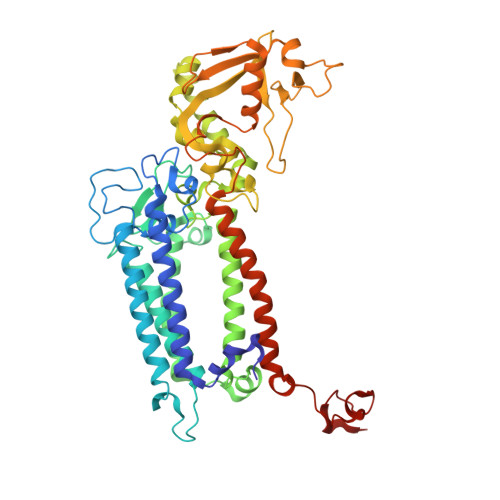
In natural photosynthesis, the light-driven splitting of water into electrons, protons and molecular oxygen forms the first step of the solar-to-chemical energy conversion process. The reaction takes place in photosystem II, where the Mn 4 CaO 5 cluster first stores four oxidizing equivalents, the S 0 to S 4 intermediate states in the Kok cycle, sequentially generated by photochemical charge separations in the reaction center and then catalyzes the O-O bond formation chemistry 1-3 . Here, we report room temperature snapshots by serial femtosecond X-ray crystallography to provide structural insights into the final reaction step of Kok's photosynthetic water oxidation cycle, the S 3 →[S 4 ]→S 0 transition where O 2 is formed and Kok's water oxidation clock is reset. Our data reveal a complex sequence of events, which occur over micro- to milliseconds, comprising changes at the Mn 4 CaO 5 cluster, its ligands and water pathways as well as controlled proton release through the hydrogen-bonding network of the Cl1 channel. Importantly, the extra O atom O x , which was introduced as a bridging ligand between Ca and Mn1 during the S 2 →S 3 transition 4-6 , disappears or relocates in parallel with Y z reduction starting at approximately 700 μs after the third flash. The onset of O 2 evolution, as indicated by the shortening of the Mn1-Mn4 distance, occurs at around 1,200 μs, signifying the presence of a reduced intermediate, possibly a bound peroxide.
Organizational Affiliation:
Molecular Biophysics and Integrated Bioimaging Division, Lawrence Berkeley National Laboratory, Berkeley, CA, USA.