Henipavirus Matrix Protein Employs a Non-Classical Nuclear Localization Signal Binding Mechanism.
Donnelly, C.M., Vogel, O.A., Edwards, M.R., Taylor, P.E., Roby, J.A., Forwood, J.K., Basler, C.F.(2023) Viruses 15
- PubMed: 37376602
- DOI: https://doi.org/10.3390/v15061302
- Primary Citation of Related Structures:
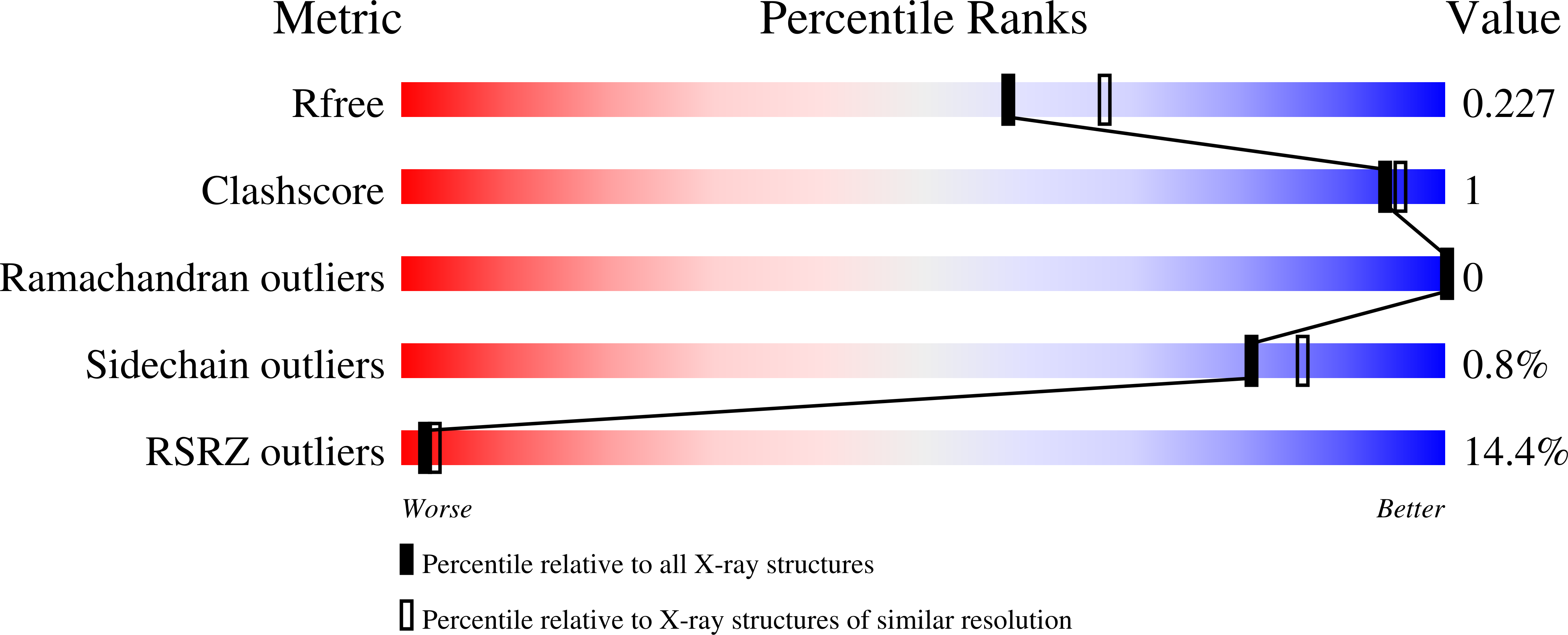
8FUA, 8FUB, 8FUC - PubMed Abstract:
Nipah virus (NiV) and Hendra virus (HeV) are highly pathogenic species from the Henipavirus genus within the paramyxovirus family and are harbored by Pteropus Flying Fox species. Henipaviruses cause severe respiratory disease, neural symptoms, and encephalitis in various animals and humans, with human mortality rates exceeding 70% in some NiV outbreaks. The henipavirus matrix protein (M), which drives viral assembly and budding of the virion, also performs non-structural functions as a type I interferon antagonist. Interestingly, M also undergoes nuclear trafficking that mediates critical monoubiquitination for downstream cell sorting, membrane association, and budding processes. Based on the NiV and HeV M X-ray crystal structures and cell-based assays, M possesses a putative monopartite nuclear localization signal (NLS) (residues 82 KRKKIR 87 ; NLS1 HeV), positioned on an exposed flexible loop and typical of how many NLSs bind importin alpha (IMPα), and a putative bipartite NLS ( 244 RR-10X-KRK 258 ; NLS2 HeV), positioned within an α-helix that is far less typical. Here, we employed X-ray crystallography to determine the binding interface of these M NLSs and IMPα. The interaction of both NLS peptides with IMPα was established, with NLS1 binding the IMPα major binding site, and NLS2 binding as a non-classical NLS to the minor site. Co-immunoprecipitation (co-IP) and immunofluorescence assays (IFA) confirm the critical role of NLS2, and specifically K258. Additionally, localization studies demonstrated a supportive role for NLS1 in M nuclear localization. These studies provide additional insight into the critical mechanisms of M nucleocytoplasmic transport, the study of which can provide a greater understanding of viral pathogenesis and uncover a potential target for novel therapeutics for henipaviral diseases.
Organizational Affiliation:
School of Dentistry and Medical Sciences, Charles Sturt University, Wagga Wagga, NSW 2678, Australia.