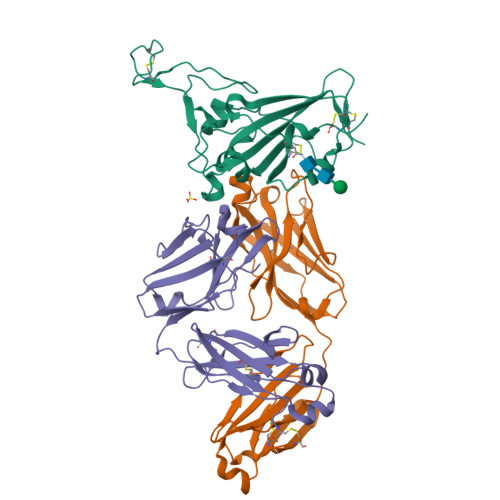
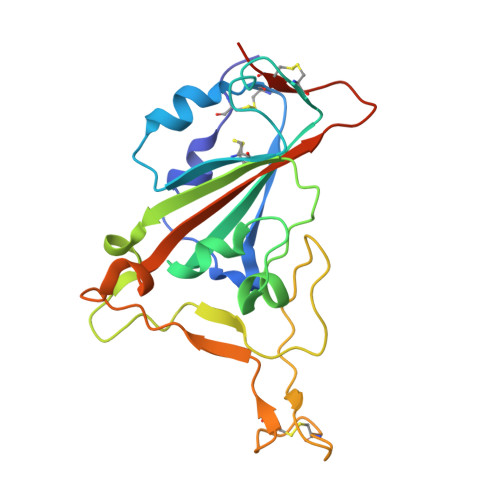
Broadly neutralizing antibodies against sarbecoviruses generated by immunization of macaques with an AS03-adjuvanted COVID-19 vaccine.
Feng, Y., Yuan, M., Powers, J.M., Hu, M., Munt, J.E., Arunachalam, P.S., Leist, S.R., Bellusci, L., Kim, J., Sprouse, K.R., Adams, L.E., Sundaramurthy, S., Zhu, X., Shirreff, L.M., Mallory, M.L., Scobey, T.D., Moreno, A., O'Hagan, D.T., Kleanthous, H., Villinger, F.J., Veesler, D., King, N.P., Suthar, M.S., Khurana, S., Baric, R.S., Wilson, I.A., Pulendran, B.(2023) Sci Transl Med 15: eadg7404-eadg7404
- PubMed: 37163615
- DOI: https://doi.org/10.1126/scitranslmed.adg7404
- Primary Citation of Related Structures:
8GB5, 8GB6, 8GB7, 8GB8 - PubMed Abstract:
The rapid emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants that evade immunity elicited by vaccination has placed an imperative on the development of countermeasures that provide broad protection against SARS-CoV-2 and related sarbecoviruses. Here, we identified extremely potent monoclonal antibodies (mAbs) that neutralized multiple sarbecoviruses from macaques vaccinated with AS03-adjuvanted monovalent subunit vaccines. Longitudinal analysis revealed progressive accumulation of somatic mutation in the immunoglobulin genes of antigen-specific memory B cells (MBCs) for at least 1 year after primary vaccination. Antibodies generated from these antigen-specific MBCs at 5 to 12 months after vaccination displayed greater potency and breadth relative to those identified at 1.4 months. Fifteen of the 338 (about 4.4%) antibodies isolated at 1.4 to 6 months after the primary vaccination showed potency against SARS-CoV-2 BA.1, despite the absence of serum BA.1 neutralization. 25F9 and 20A7 neutralized authentic clade 1 sarbecoviruses (SARS-CoV, WIV-1, SHC014, SARS-CoV-2 D614G, BA.1, and Pangolin-GD) and vesicular stomatitis virus-pseudotyped clade 3 sarbecoviruses (BtKY72 and PRD-0038). 20A7 and 27A12 showed potent neutralization against all SARS-CoV-2 variants and multiple Omicron sublineages, including BA.1, BA.2, BA.3, BA.4/5, BQ.1, BQ.1.1, and XBB. Crystallography studies revealed the molecular basis of broad and potent neutralization through targeting conserved sites within the RBD. Prophylactic protection of 25F9, 20A7, and 27A12 was confirmed in mice, and administration of 25F9 particularly provided complete protection against SARS-CoV-2, BA.1, SARS-CoV, and SHC014 challenge. These data underscore the extremely potent and broad activity of these mAbs against sarbecoviruses.
Organizational Affiliation:
Institute for Immunity, Transplantation and Infection, Stanford University, Stanford, CA 94305, USA.