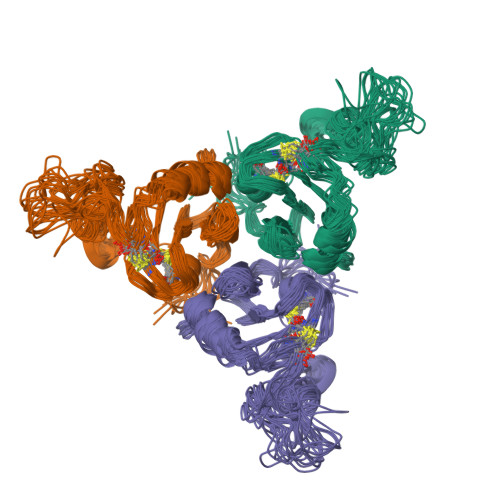
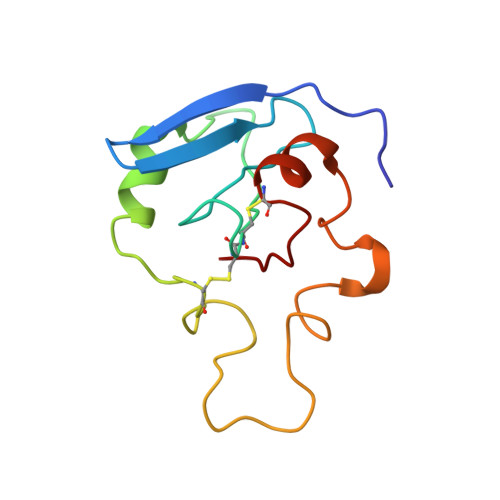
The inhibitory action of the chaperone BRICHOS against the alpha-Synuclein secondary nucleation pathway.
Ghosh, D., Torres, F., Schneider, M.M., Ashkinadze, D., Kadavath, H., Fleischmann, Y., Mergenthal, S., Guntert, P., Krainer, G., Andrzejewska, E.A., Lin, L., Wei, J., Klotzsch, E., Knowles, T., Riek, R.(2024) Nat Commun 15: 10038-10038
- PubMed: 39567476
- DOI: https://doi.org/10.1038/s41467-024-54212-2
- Primary Citation of Related Structures:
8OVI, 8OX2 - PubMed Abstract:
The complex kinetics of disease-related amyloid aggregation of proteins such as α-Synuclein (α-Syn) in Parkinson's disease and Aβ42 in Alzheimer's disease include primary nucleation, amyloid fibril elongation and secondary nucleation. The latter can be a key accelerator of the aggregation process. It has been demonstrated that the chaperone domain BRICHOS can interfere with the secondary nucleation process of Aβ42. Here, we explore the mechanism of secondary nucleation inhibition of the BRICHOS domain of the lung surfactant protein (proSP-C) against α-Syn aggregation and amyloid formation. We determine the 3D NMR structure of an inactive trimer of proSP-C BRICHOS and its active monomer using a designed mutant. Furthermore, the interaction between the proSP-C BRICHOS chaperone and a substrate peptide has been studied. NMR-based interaction studies of proSP-C BRICHOS with α-Syn fibrils show that proSP-C BRICHOS binds to the C-terminal flexible fuzzy coat of the fibrils, which is the secondary nucleation site on the fibrils. Super-resolution fluorescence microscopy demonstrates that proSP-C BRICHOS runs along the fibrillar axis diffusion-dependently sweeping off monomeric α-Syn from the fibrils. The observed mechanism explains how a weakly binding chaperone can inhibit the α-Syn secondary nucleation pathway via avidity where a single proSP-C BRICHOS molecule is sufficient against up to ~7-40 α-Syn molecules embedded within the fibrils.
Organizational Affiliation:
Institute of Molecular Physical Science (IMPS), ETH Zürich, Vladimir-Prelog-Weg 2, CH-8093, Zürich, Switzerland.