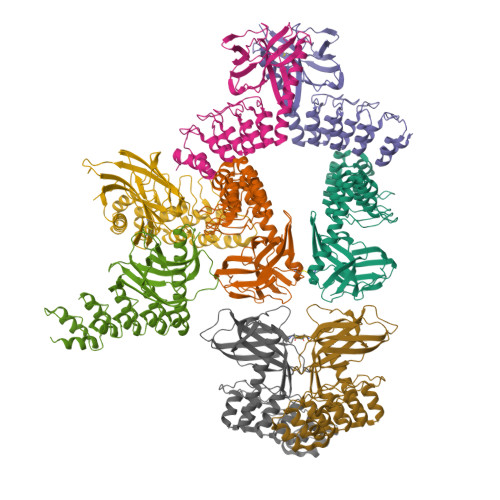
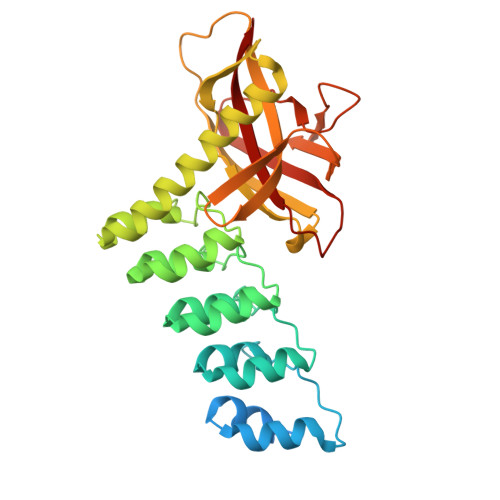
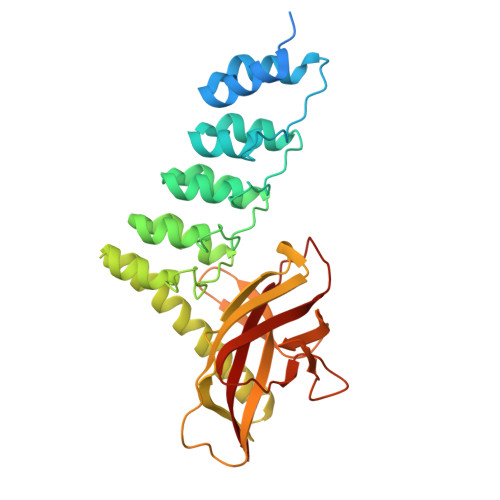
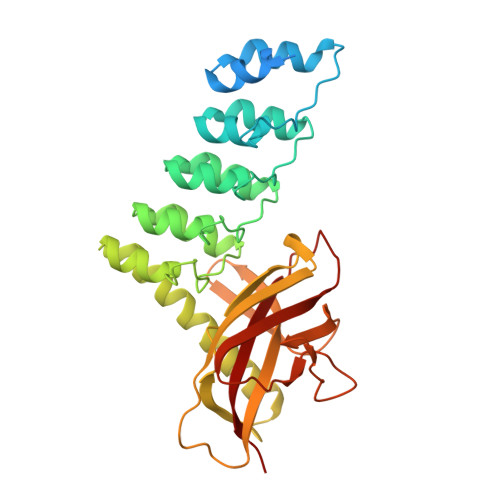
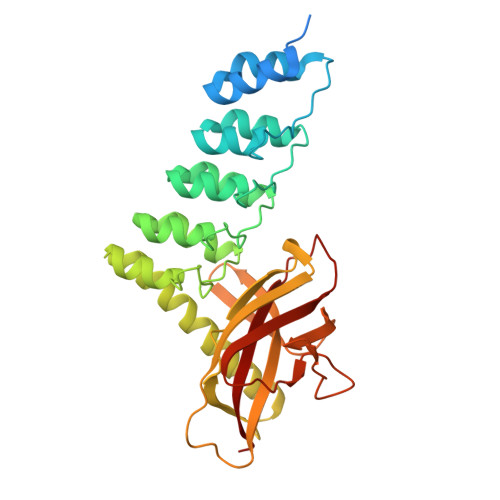
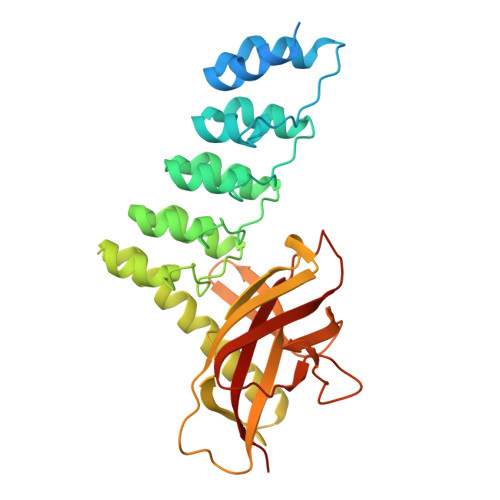
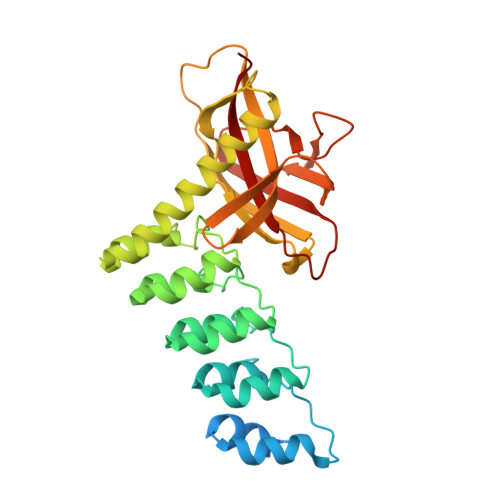
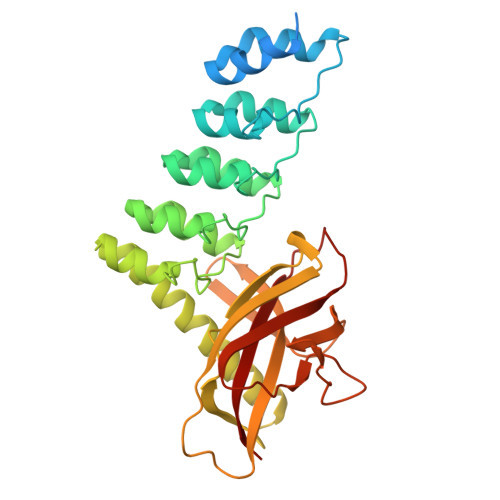
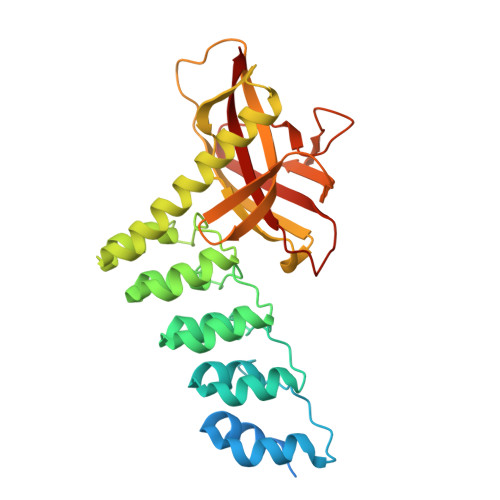
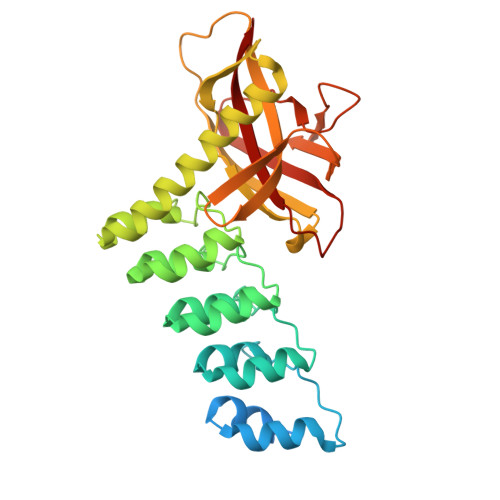
Structure and Methyl-lysine Binding Selectivity of the HUSH Complex Subunit MPP8.
Nikolopoulos, N., Oda, S.I., Prigozhin, D.M., Modis, Y.(2024) J Mol Biology 437: 168890-168890
- PubMed: 39638237
- DOI: https://doi.org/10.1016/j.jmb.2024.168890
- Primary Citation of Related Structures:
8QFB - PubMed Abstract:
The Human Silencing Hub (HUSH) guards the genome from the pathogenic effects of retroelement expression. Composed of MPP8, TASOR, and Periphilin-1, HUSH recognizes actively transcribed retrotransposed sequences by the presence of long (>1.5-kb) nascent transcripts without introns. HUSH recruits effectors that alter chromatin structure, degrade transcripts, and deposit transcriptionally repressive epigenetic marks. Here, we report the crystal structure of the C-terminal domain (CTD) of MPP8 necessary for HUSH activity. The MPP8 CTD consists of five ankyrin repeats followed by a domain with structural homology to the PINIT domains of Siz/PIAS-family SUMO E3 ligases. AlphaFold3 modeling of the MPP8-TASOR complex predicts that a SPOC domain and a domain with a novel fold in TASOR form extended interaction interfaces with the MPP8 CTD. Point mutations at these interfaces resulted in loss of HUSH-dependent transcriptional repression in a cell-based reporter assay, validating the AlphaFold3 model. The MPP8 chromodomain, known to bind the repressive mark H3K9me3, bound with similar or higher affinity to sequences in the H3K9 methyltransferase subunits SETDB1, ATF7IP, G9a, and GLP. Hence, MPP8 promotes heterochromatinization by recruiting H3K9 methyltransferases. Our work identifies novel structural elements in MPP8 required for HUSH complex assembly and silencing, thereby fulfilling vital functions in controlling retrotransposons.
Organizational Affiliation:
Molecular Immunity Unit, Department of Medicine, University of Cambridge, MRC Laboratory of Molecular Biology, Cambridge CB2 0QH, UK; Cambridge Institute of Therapeutic Immunology & Infectious Disease (CITIID), Department of Medicine, University of Cambridge, Cambridge CB2 0AW, UK.