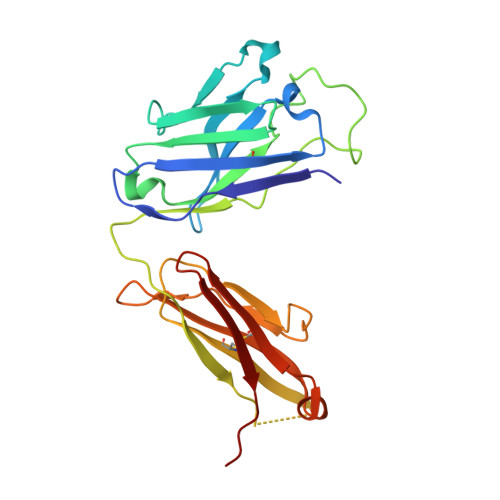
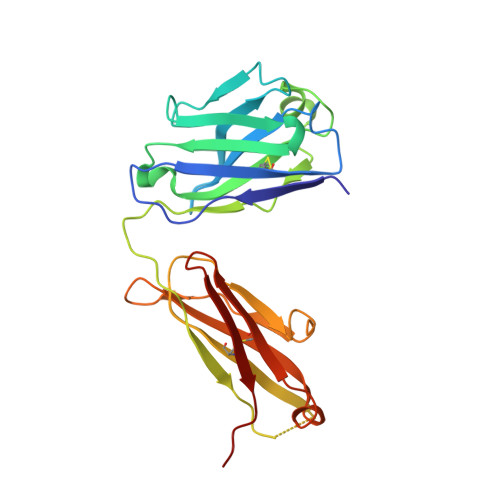
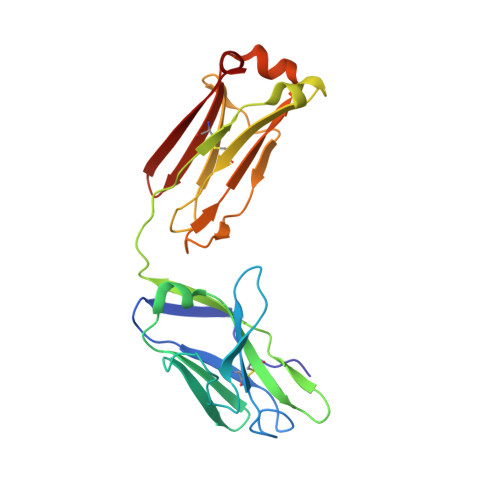
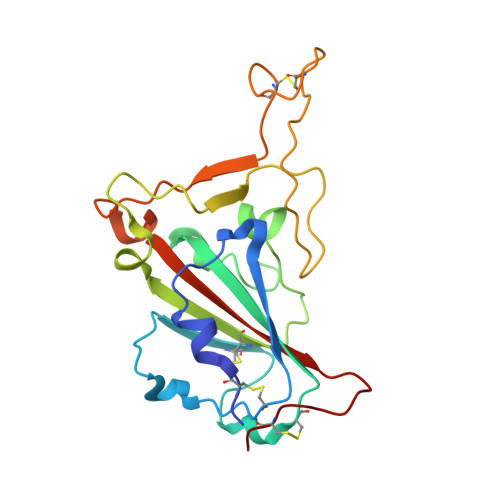
AZD3152 neutralizes SARS-CoV-2 historical and contemporary variants and is protective in hamsters and well tolerated in adults.
Cai, Y., Diallo, S., Rosenthal, K., Ren, K., Flores, D.J., Dippel, A., Oganesyan, V., van Dyk, N., Chen, X., Cantu, E., Choudhary, R., Sulikowski, M., Adissu, H., Chawla, B., Kar, S., Liu, C., Dijokaite-Guraliuc, A., Mongkolsapaya, J., Rajan, S., Loo, Y.M., Beavon, R., Webber, C., Chang, L.J., Thomas, S., Clegg, L., Zhang, H., Screaton, G.R., Philbin, N., Harre, M., Selim, A., Martinez-Alier, N., Uriel, A., Cohen, T.S., Perez, J.L., Esser, M.T., Blair, W., Francica, J.R.(2024) Sci Transl Med 16: eado2817-eado2817
- PubMed: 38924429
- DOI: https://doi.org/10.1126/scitranslmed.ado2817
- Primary Citation of Related Structures:
8SUO - PubMed Abstract:
The evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in variants that can escape neutralization by therapeutic antibodies. Here, we describe AZD3152, a SARS-CoV-2-neutralizing monoclonal antibody designed to provide improved potency and coverage against emerging variants. AZD3152 binds to the back left shoulder of the SARS-CoV-2 spike protein receptor binding domain and prevents interaction with the human angiotensin-converting enzyme 2 receptor. AZD3152 potently neutralized a broad panel of pseudovirus variants, including the currently dominant Omicron variant JN.1 but has reduced potency against XBB subvariants containing F456L. In vitro studies confirmed F456L resistance and additionally identified T415I and K458E as escape mutations. In a Syrian hamster challenge model, prophylactic administration of AZD3152 protected hamsters from weight loss and inflammation-related lung pathologies and reduced lung viral load. In the phase 1 sentinel safety cohort of the ongoing SUPERNOVA study (ClinicalTrials.gov: NCT05648110), a single 600-mg intramuscular injection of AZD5156 (containing 300 mg each of AZD3152 and cilgavimab) was well tolerated in adults through day 91. Observed serum concentrations of AZD3152 through day 91 were similar to those observed with cilgavimab and consistent with predictions for AZD7442, a SARS-CoV-2-neutralizing antibody combination of cilgavimab and tixagevimab, in a population pharmacokinetic model. On the basis of its pharmacokinetic characteristics, AZD3152 is predicted to provide durable protection against symptomatic coronavirus disease 2019 caused by susceptible SARS-CoV-2 variants, such as JN.1, in humans.
Organizational Affiliation:
Vaccines and Immune Therapies, BioPharmaceuticals R&D, AstraZeneca, Gaithersburg, MD 20878, USA.