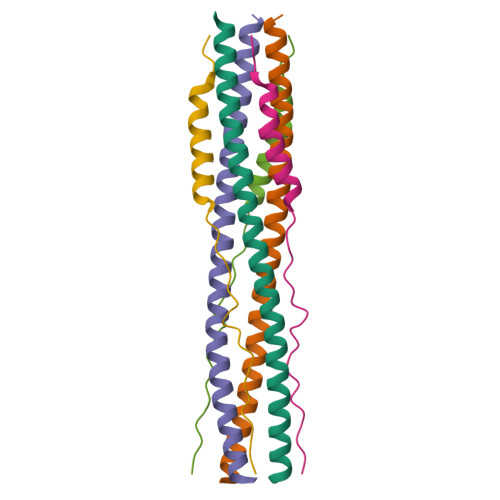
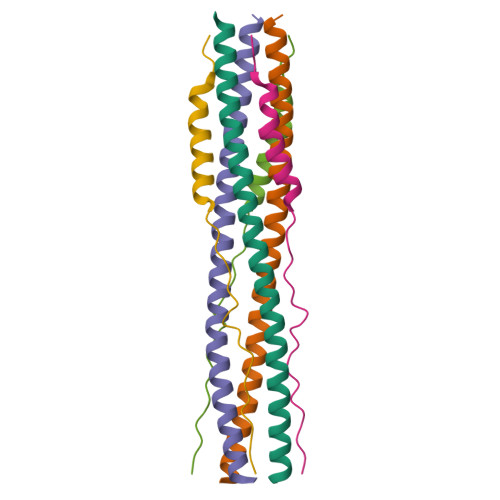
Nanomolar inhibition of SARS-CoV-2 infection by an unmodified peptide targeting the prehairpin intermediate of the spike protein.
Yang, K., Wang, C., Kreutzberger, A.J.B., Ojha, R., Kuivanen, S., Couoh-Cardel, S., Muratcioglu, S., Eisen, T.J., White, K.I., Held, R.G., Subramanian, S., Marcus, K., Pfuetzner, R.A., Esquivies, L., Doyle, C.A., Kuriyan, J., Vapalahti, O., Balistreri, G., Kirchhausen, T., Brunger, A.T.(2022) Proc Natl Acad Sci U S A 119: e2210990119-e2210990119
- PubMed: 36122200
- DOI: https://doi.org/10.1073/pnas.2210990119
- Primary Citation of Related Structures:
8CZI - PubMed Abstract:
Variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) challenge currently available coronavirus disease 2019 vaccines and monoclonal antibody therapies through epitope change on the receptor binding domain of the viral spike glycoprotein. Hence, there is a specific urgent need for alternative antivirals that target processes less likely to be affected by mutation, such as the membrane fusion step of viral entry into the host cell. One such antiviral class includes peptide inhibitors, which block formation of the so-called heptad repeat 1 and 2 (HR1HR2) six-helix bundle of the SARS-CoV-2 spike (S) protein and thus interfere with viral membrane fusion. We performed structural studies of the HR1HR2 bundle, revealing an extended, well-folded N-terminal region of HR2 that interacts with the HR1 triple helix. Based on this structure, we designed an extended HR2 peptide that achieves single-digit nanomolar inhibition of SARS-CoV-2 in cell-based and virus-based assays without the need for modifications such as lipidation or chemical stapling. The peptide also strongly inhibits all major SARS-CoV-2 variants to date. This extended peptide is ∼100-fold more potent than all previously published short, unmodified HR2 peptides, and it has a very long inhibition lifetime after washout in virus infection assays, suggesting that it targets a prehairpin intermediate of the SARS-CoV-2 S protein. Together, these results suggest that regions outside the HR2 helical region may offer new opportunities for potent peptide-derived therapeutics for SARS-CoV-2 and its variants, and even more distantly related viruses, and provide further support for the prehairpin intermediate of the S protein.
Organizational Affiliation:
HHMI, Stanford University, Stanford, CA 94305.