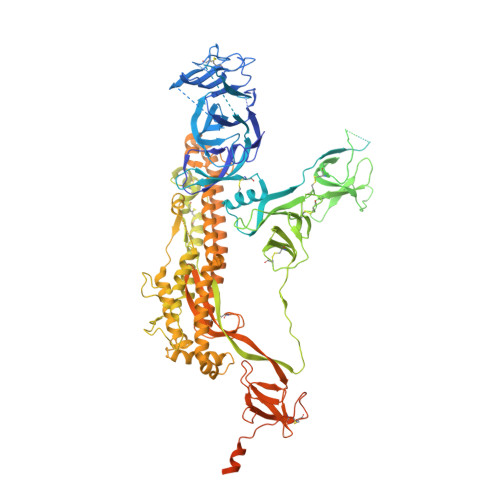
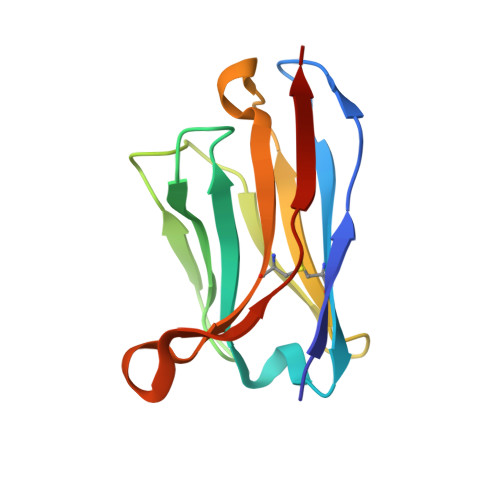
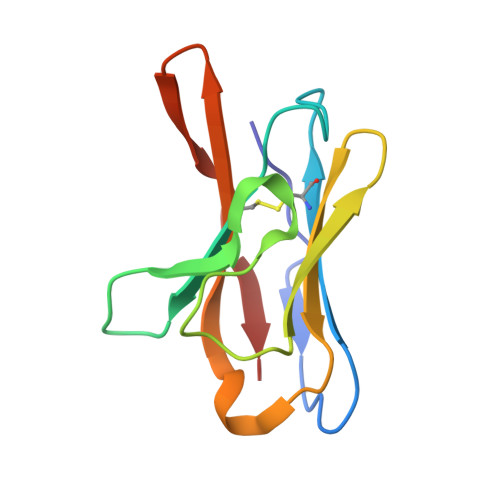
Site of vulnerability on SARS-CoV-2 spike induces broadly protective antibody against antigenically distinct Omicron subvariants.
Changrob, S., Halfmann, P.J., Liu, H., Torres, J.L., McGrath, J.J.C., Ozorowski, G., Li, L., Wilbanks, G.D., Kuroda, M., Maemura, T., Huang, M., Zheng, N.Y., Turner, H.L., Erickson, S.A., Fu, Y., Yasuhara, A., Singh, G., Monahan, B., Mauldin, J., Srivastava, K., Simon, V., Krammer, F., Sather, D.N., Ward, A.B., Wilson, I.A., Kawaoka, Y., Wilson, P.C.(2023) J Clin Invest 133
- PubMed: 36862518
- DOI: https://doi.org/10.1172/JCI166844
- Primary Citation of Related Structures:
8D0Z - PubMed Abstract:
The rapid evolution of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron variants has emphasized the need to identify antibodies with broad neutralizing capabilities to inform future monoclonal therapies and vaccination strategies. Herein, we identified S728-1157, a broadly neutralizing antibody (bnAb) targeting the receptor-binding site (RBS) that was derived from an individual previously infected with WT SARS-CoV-2 prior to the spread of variants of concern (VOCs). S728-1157 demonstrated broad cross-neutralization of all dominant variants, including D614G, Beta, Delta, Kappa, Mu, and Omicron (BA.1/BA.2/BA.2.75/BA.4/BA.5/BL.1/XBB). Furthermore, S728-1157 protected hamsters against in vivo challenges with WT, Delta, and BA.1 viruses. Structural analysis showed that this antibody targets a class 1/RBS-A epitope in the receptor binding domain via multiple hydrophobic and polar interactions with its heavy chain complementarity determining region 3 (CDR-H3), in addition to common motifs in CDR-H1/CDR-H2 of class 1/RBS-A antibodies. Importantly, this epitope was more readily accessible in the open and prefusion state, or in the hexaproline (6P)-stabilized spike constructs, as compared with diproline (2P) constructs. Overall, S728-1157 demonstrates broad therapeutic potential and may inform target-driven vaccine designs against future SARS-CoV-2 variants.
Organizational Affiliation:
Drukier Institute for Children's Health, Department of Pediatrics, Weill Cornell Medicine, New York, New York, USA.