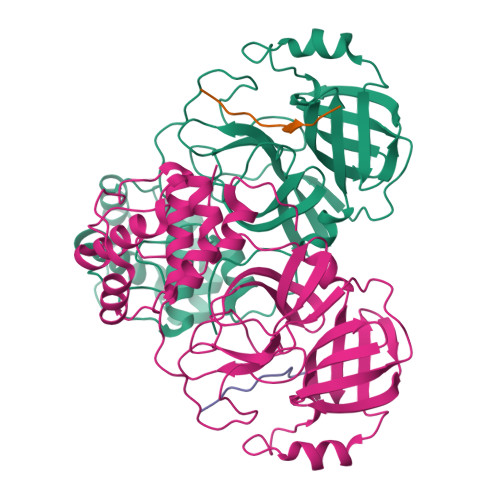
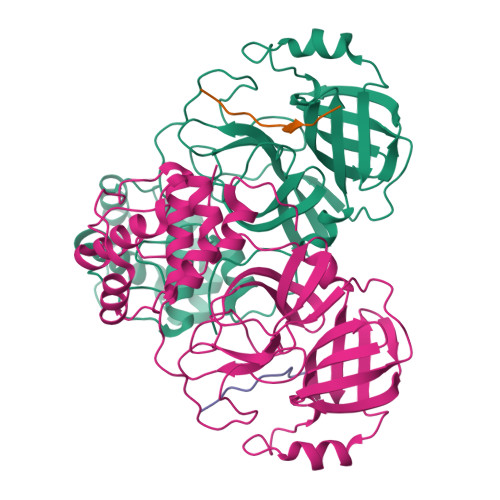
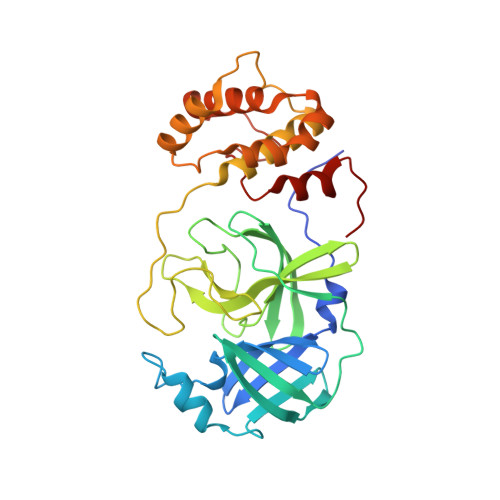
An in-solution snapshot of SARS-COV-2 main protease maturation process and inhibition.
Noske, G.D., Song, Y., Fernandes, R.S., Chalk, R., Elmassoudi, H., Koekemoer, L., Owen, C.D., El-Baba, T.J., Robinson, C.V., Oliva, G., Godoy, A.S.(2023) Nat Commun 14: 1545-1545
- PubMed: 36941262
- DOI: https://doi.org/10.1038/s41467-023-37035-5
- Primary Citation of Related Structures:
8EY2, 8EYJ - PubMed Abstract:
The main protease from SARS-CoV-2 (M pro ) is responsible for cleavage of the viral polyprotein. M pro self-processing is called maturation, and it is crucial for enzyme dimerization and activity. Here we use C145S M pro to study the structure and dynamics of N-terminal cleavage in solution. Native mass spectroscopy analysis shows that mixed oligomeric states are composed of cleaved and uncleaved particles, indicating that N-terminal processing is not critical for dimerization. A 3.5 Å cryo-EM structure provides details of M pro N-terminal cleavage outside the constrains of crystal environment. We show that different classes of inhibitors shift the balance between oligomeric states. While non-covalent inhibitor MAT-POS-e194df51-1 prevents dimerization, the covalent inhibitor nirmatrelvir induces the conversion of monomers into dimers, even with intact N-termini. Our data indicates that the M pro dimerization is triggered by induced fit due to covalent linkage during substrate processing rather than the N-terminal processing.
Organizational Affiliation:
Sao Carlos Institute of Physics, University of Sao Paulo, Av. Joao Dagnone, 1100 - Jardim Santa Angelina, Sao Carlos, 13563-120, Brazil.