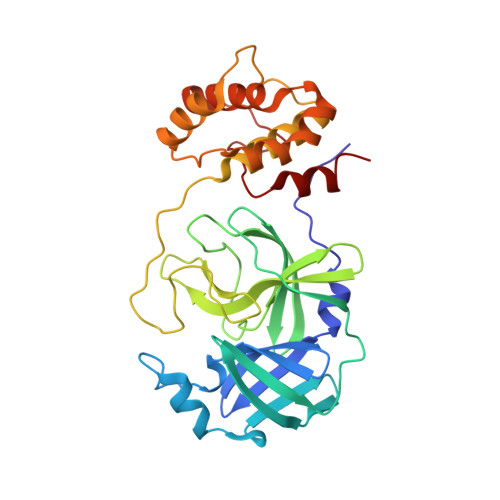
The S1'-S3' Pocket of the SARS-CoV-2 Main Protease Is Critical for Substrate Selectivity and Can Be Targeted with Covalent Inhibitors.
Liu, M., Li, J., Liu, W., Yang, Y., Zhang, M., Ye, Y., Zhu, W., Zhou, C., Zhai, H., Xu, Z., Zhang, G., Huang, H.(2023) Angew Chem Int Ed Engl 62: e202309657-e202309657
- PubMed: 37609788
- DOI: https://doi.org/10.1002/anie.202309657
- Primary Citation of Related Structures:
8GW4, 8GWS, 8JPQ - PubMed Abstract:
The main protease (M pro ) of SARS-CoV-2 is a well-characterized target for antiviral drug discovery. To date, most antiviral drug discovery efforts have focused on the S4-S1' pocket of M pro ; however, it is still unclear whether the S1'-S3' pocket per se can serve as a new site for drug discovery. In this study, the S1'-S3' pocket of M pro was found to differentially recognize viral peptidyl substrates. For instance, S3' in M pro strongly favors Phe or Trp, and S1' favors Ala. The peptidyl inhibitor D-4-77, which possesses an α-bromoacetamide warhead, was discovered to be a promising inhibitor of M pro , with an IC 50 of 0.95 μM and an antiviral EC 50 of 0.49 μM. The M pro /inhibitor co-crystal structure confirmed the binding mode of the inhibitor to the S1'-S3' pocket and revealed a covalent mechanism. In addition, D-4-77 functions as an immune protectant and suppresses SARS-CoV-2 M pro -induced antagonism of the host NF-κB innate immune response. These findings indicate that the S1'-S3' pocket of SARS-CoV-2 M pro is druggable, and that inhibiting SARS-CoV-2 M pro can simultaneously protect human innate immunity and inhibit virion assembly.
Organizational Affiliation:
State Key Laboratory of Chemical Oncogenomics, Guangdong Provincial Key Laboratory of Chemical Genomics, Laboratory of Structural Biology and Drug Discovery, Laboratory of Ubiquitination and Targeted Therapy, School of Chemical Biology and Biotechnology, Peking University Shenzhen Graduate School, Shenzhen, Guangdong, 518055, China.