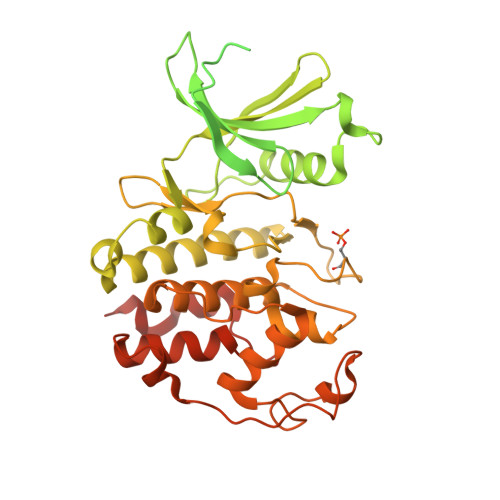
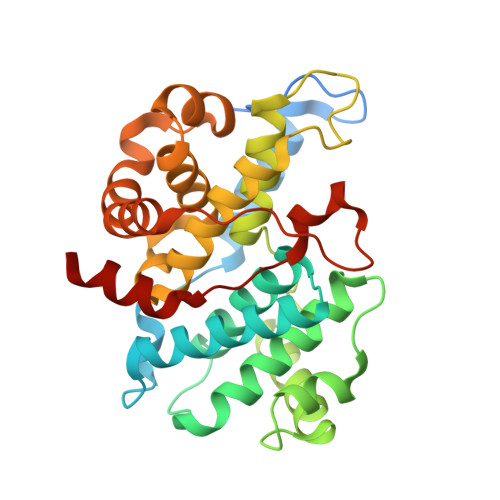
Structural basis of CDK3 activation by cyclin E1 and inhibition by dinaciclib.
Gui, W., Hang, Y., Cheng, W., Gao, M., Wu, J., Ouyang, Z.(2023) Biochem Biophys Res Commun 662: 126-134
- PubMed: 37104883
- DOI: https://doi.org/10.1016/j.bbrc.2023.04.026
- Primary Citation of Related Structures:
7XQK, 8H4R - PubMed Abstract:
Cell cycle transitions are controlled by multiple cell cycle regulators, especially CDKs. Several CDKs, including CDK1-4 and CDK6, promote cell cycle progression directly. Among them, CDK3 is critically important because it triggers the transitions of G0 to G1 and G1 to S phase through binding to cyclin C and cyclin E1, respectively. In contrast to its highly related homologs, the molecular basis of CDK3 activation remains elusive due to the lack of structural information of CDK3, particularly in cyclin bound form. Here we report the crystal structure of CDK3 in complex with cyclin E1 at 2.25 Å resolution. CDK3 resembles CDK2 in that both adopt a similar fold and bind cyclin E1 in a similar way. The structural discrepancy between CDK3 and CDK2 may reflect their substrate specificity. Profiling a panel of CDK inhibitors reveals that dinaciclib inhibits CDK3-cyclin E1 potently and specifically. The structure of CDK3-cyclin E1 bound to dinaciclib reveals the inhibitory mechanism. The structural and biochemical results uncover the mechanism of CDK3 activation by cyclin E1 and lays a foundation for structural-based drug design.
Organizational Affiliation:
Wuxi Biortus Biosciences Co. Ltd, 6 Dongsheng Western Road, Jiangyin, Jiangsu, 214437, China.