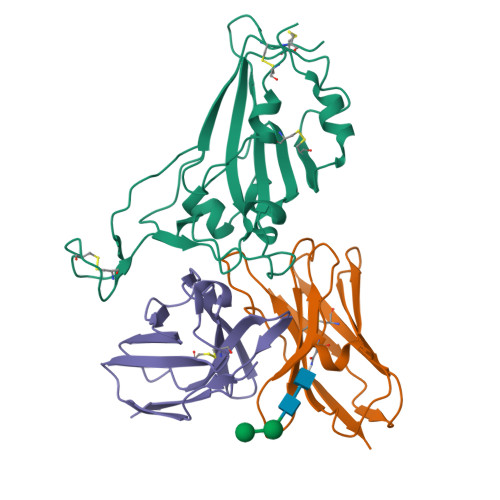
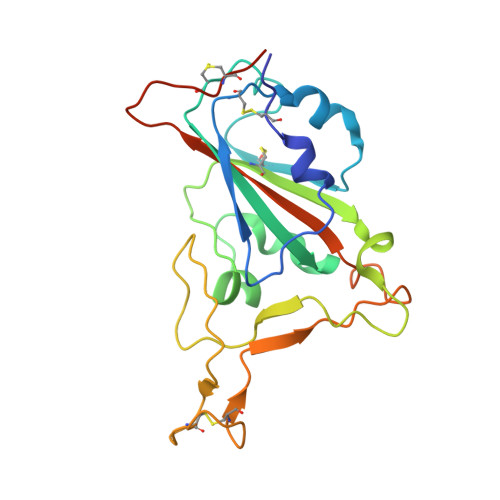
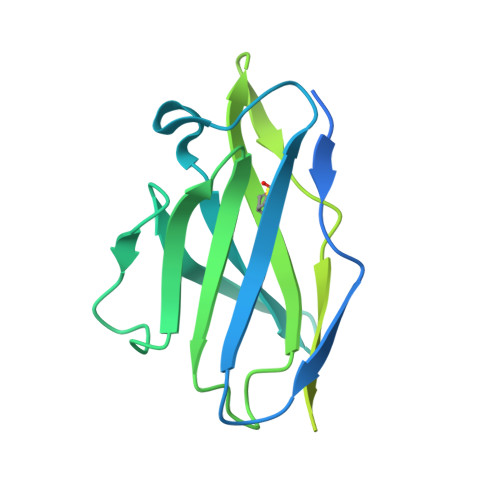
The structure of the RBD-E77 Fab complex reveals neutralization and immune escape of SARS-CoV-2.
Zhang, Z., Li, X., Xue, Y., Yang, B., Jia, Y., Liu, S., Lu, D.(2023) Acta Crystallogr D Struct Biol 79: 746-757
- PubMed: 37428848
- DOI: https://doi.org/10.1107/S2059798323005041
- Primary Citation of Related Structures:
8IDN - PubMed Abstract:
The spike protein (S) of SARS-CoV-2 is the major target of neutralizing antibodies and vaccines. Antibodies that target the receptor-binding domain (RBD) of S have high potency in preventing viral infection. The ongoing evolution of SARS-CoV-2, especially mutations occurring in the RBD of new variants, has severely challenged the development of neutralizing antibodies and vaccines. Here, a murine monoclonal antibody (mAb) designated E77 is reported which engages the prototype RBD with high affinity and potently neutralizes SARS-CoV-2 pseudoviruses. However, the capability of E77 to bind RBDs vanishes upon encountering variants of concern (VOCs) which carry the N501Y mutation, such as Alpha, Beta, Gamma and Omicron, in contrast to its performance with the Delta variant. To explain the discrepancy, cryo-electron microscopy was used to analyze the structure of an RBD-E77 Fab complex, which reveals that the binding site of E77 on RBD belongs to the RBD-1 epitope, which largely overlaps with the binding site of human angiotensin-converting enzyme 2 (hACE2). Both the heavy chain and the light chain of E77 interact extensively with RBD and contribute to the strong binding of RBD. E77 employs CDRL1 to engage Asn501 of RBD and the Asn-to-Tyr mutation could generate steric hindrance, abolishing the binding. In sum, the data provide the landscape for an in-depth understanding of immune escape of VOCs and rational antibody engineering against emerging variants of SARS-CoV-2.
Organizational Affiliation:
College of Life Sciences, Shanxi Agricultural University, Taiyuan 030031, People's Republic of China.