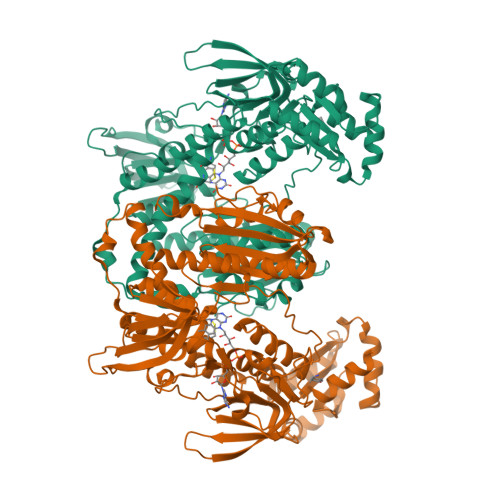
Fragment library screening by X-ray crystallography and binding site analysis on thioredoxin glutathione reductase of Schistosoma mansoni.
de Souza Neto, L.R., Montoya, B.O., Brandao-Neto, J., Verma, A., Bowyer, S., Moreira-Filho, J.T., Dantas, R.F., Neves, B.J., Andrade, C.H., von Delft, F., Owens, R.J., Furnham, N., Silva-Jr, F.P.(2024) Sci Rep 14: 1582-1582
- PubMed: 38238498
- DOI: https://doi.org/10.1038/s41598-024-52018-2
- Primary Citation of Related Structures:
8PDD, 8PL0, 8PL1, 8PL2, 8PL3, 8PL4, 8PL5, 8PL6, 8PL7, 8PL8, 8PL9, 8PLA, 8PLB, 8PLC, 8PLD, 8PLE, 8PLF, 8PLG, 8PLH, 8PLI, 8PLJ, 8PLK, 8PLL, 8PLM, 8PLN, 8PLO, 8PLP, 8PLQ, 8PLR, 8PLS, 8PLT, 8PLU, 8PLV, 8PLW, 8PLX, 8PLY - PubMed Abstract:
Schistosomiasis is caused by parasites of the genus Schistosoma, which infect more than 200 million people. Praziquantel (PZQ) has been the main drug for controlling schistosomiasis for over four decades, but despite that it is ineffective against juvenile worms and size and taste issues with its pharmaceutical forms impose challenges for treating school-aged children. It is also important to note that PZQ resistant strains can be generated in laboratory conditions and observed in the field, hence its extensive use in mass drug administration programs raises concerns about resistance, highlighting the need to search for new schistosomicidal drugs. Schistosomes survival relies on the redox enzyme thioredoxin glutathione reductase (TGR), a validated target for the development of new anti-schistosomal drugs. Here we report a high-throughput fragment screening campaign of 768 compounds against S. mansoni TGR (SmTGR) using X-ray crystallography. We observed 49 binding events involving 35 distinct molecular fragments which were found to be distributed across 16 binding sites. Most sites are described for the first time within SmTGR, a noteworthy exception being the "doorstop pocket" near the NADPH binding site. We have compared results from hotspots and pocket druggability analysis of SmTGR with the experimental binding sites found in this work, with our results indicating only limited coincidence between experimental and computational results. Finally, we discuss that binding sites at the doorstop/NADPH binding site and in the SmTGR dimer interface, should be prioritized for developing SmTGR inhibitors as new antischistosomal drugs.
Organizational Affiliation:
LaBECFar - Laboratory of Experimental and Computational Biochemistry of Drugs, Oswaldo Cruz Institute, FIOCRUZ, Rio de Janeiro, Brazil.