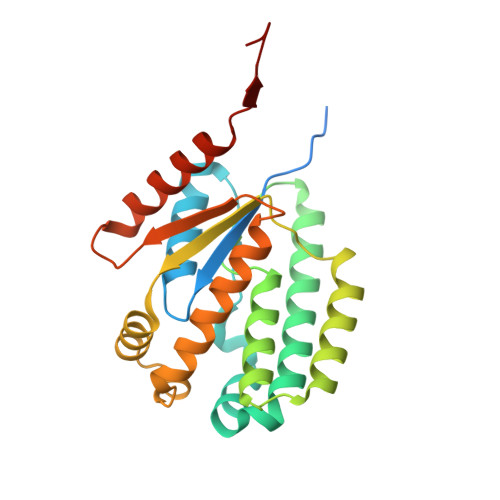
Tetramerization of deoxyadenosine kinase meets the demands of a DNA replication substrate challenge in Giardia intestinalis.
Ranjbarian, F., Rafie, K., Shankar, K., Krakovka, S., Svard, S.G., Carlson, L.A., Hofer, A.(2024) Nucleic Acids Res
- PubMed: 39607702
- DOI: https://doi.org/10.1093/nar/gkae1073
- Primary Citation of Related Structures:
8PUU - PubMed Abstract:
The protozoan parasite Giardia intestinalis is one of only a few organisms lacking de novo synthesis of DNA building blocks (deoxyribonucleotides). Instead, the parasite relies exclusively on salvaging deoxyadenosine and other deoxyribonucleosides from its host environment. Here, we report that G. intestinalis has a deoxyribonucleoside kinase with a 1000-fold higher catalytic efficiency (kcat/KM) for deoxyadenosine than the corresponding mammalian kinases and can thereby provide sufficient deoxyadenosine triphosphate levels for DNA synthesis despite the lack of de novo synthesis. Several deoxyadenosine analogs were also potent substrates and showed comparable EC50 values on cultured G. intestinalis cells as metronidazole, the current first-line treatment, with the additional advantage of being effective against metronidazole-resistant parasites. Structural analysis using cryo-EM and X-ray crystallography showed that the enzyme is unique within its family of deoxyribonucleoside kinases by forming a tetramer stabilized by extended N- and C-termini in a novel dimer-dimer interaction. Removal of the two termini resulted in lost ability to form tetramers and a markedly reduced affinity for the deoxyribonucleoside substrate. The development of highly efficient deoxyribonucleoside kinases via oligomerization may represent a critical evolutionary adaptation in organisms that rely solely on deoxyribonucleoside salvage.
Organizational Affiliation:
Department of Medical Biochemistry and Biophysics, Umeå University, Linnaeus väg 6, SE-901 87 Umeå, Sweden.