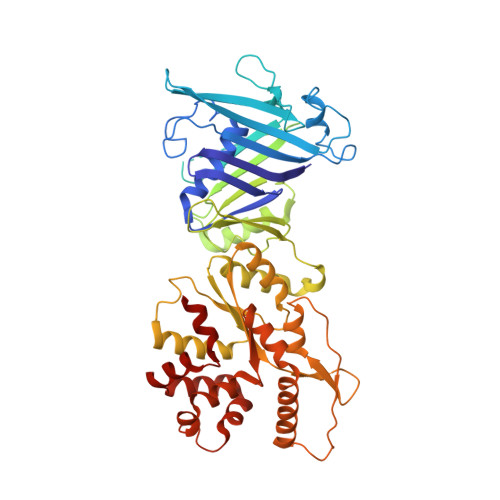
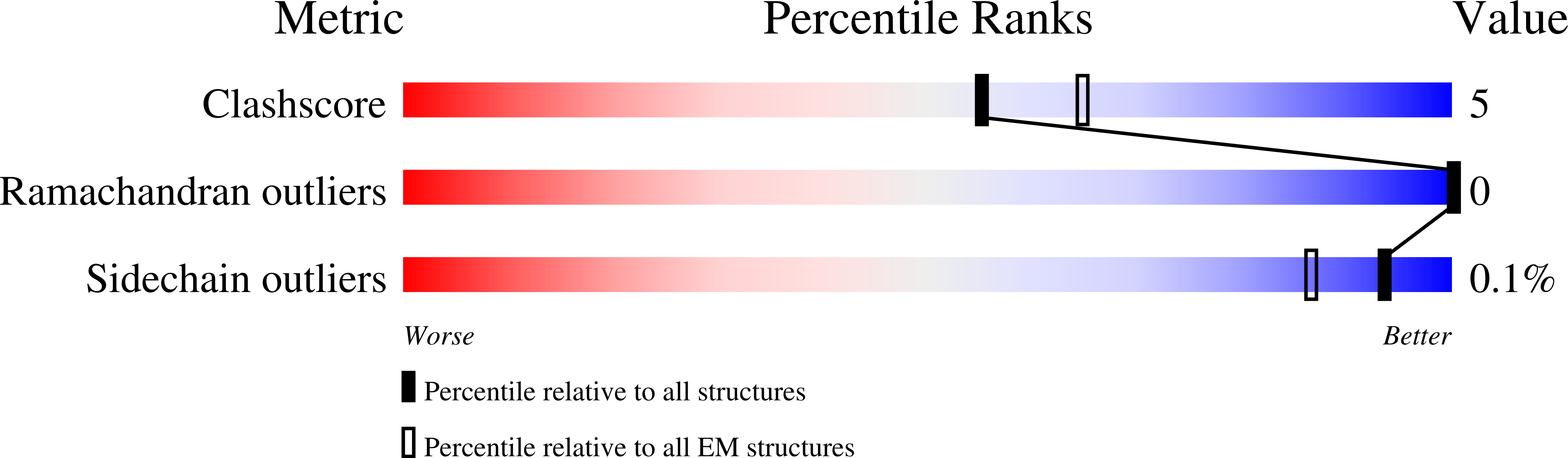Molecular basis of Gabija anti-phage supramolecular assemblies.
Yang, X.Y., Shen, Z., Xie, J., Greenwald, J., Marathe, I., Lin, Q., Xie, W.J., Wysocki, V.H., Fu, T.M.(2024) Nat Struct Mol Biol 31: 1243-1250
- PubMed: 38627580
- DOI: https://doi.org/10.1038/s41594-024-01283-w
- Primary Citation of Related Structures:
8TJY, 8TK0, 8TK1 - PubMed Abstract:
As one of the most prevalent anti-phage defense systems in prokaryotes, Gabija consists of a Gabija protein A (GajA) and a Gabija protein B (GajB). The assembly and function of the Gabija system remain unclear. Here we present cryo-EM structures of Bacillus cereus GajA and GajAB complex, revealing tetrameric and octameric assemblies, respectively. In the center of the complex, GajA assembles into a tetramer, which recruits two sets of GajB dimer at opposite sides of the complex, resulting in a 4:4 GajAB supramolecular complex for anti-phage defense. Further biochemical analysis showed that GajA alone is sufficient to cut double-stranded DNA and plasmid DNA, which can be inhibited by ATP. Unexpectedly, the GajAB displays enhanced activity for plasmid DNA, suggesting a role of substrate selection by GajB. Together, our study defines a framework for understanding anti-phage immune defense by the GajAB complex.
Organizational Affiliation:
Department of Biological Chemistry and Pharmacology, Center for RNA Biology, The Ohio State University, Columbus, OH, USA.